Abstract
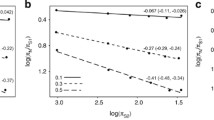
On the relationship between mitochondrion and longevity in mammals, two viewpoints have been proposed: one found that the amino acid substitution rates of most of the mitochondrial DNA-coding peptides were positively correlated with longevity, while the other raised the opposite view. To resolve this dichotomy, and to explore the relationship between mtDNA evolution and longevity in mammals, we examined this relationship in 85 mammal species, at the nucleotide sequence level. Previous studies have demonstrated that phylogenetic inertia, substitution saturation, and body mass can affect the relationship between longevity and substitution rate. Therefore, analyses should take these factors into account. This study found that after controlling the aforementioned factors, no significant positive or negative relationship existed between mitochondrial DNA evolutionary rate and longevity except for CYTB, partly agreeing with a previous study. Variations of longevity can be explained partly by the evolutionary rate of CYTB, but other influencing factors still need to be studied in the future.



Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Barja G, Herrero A (2000) Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. Faseb J 14:312–318
Bennettand PM, Harvey PH (2009) Active and resting metabolism in birds: allometry, phylogeny and ecology. J Zool 213:327–344
Bromham L, Rambaut A, Harvey PH (1996) Determinants of rate variation in mammalian DNA sequence evolution. J Mol Evol 43:610–621
Brown-Borg HM (2007) Hormonal regulation of longevity in mammals. Ageing Res Rev 6:28–45.
Capellini I, Venditti C, Barton R (2010) Phylogeny and metabolic scaling in mammals. Ecology 91:2783–2793
Crofts AR, Lhee S, Crofts SB,Cheng J, Rose S (2006) Proton pumping in the bc1 complex: a new gating mechanism that prevents short circuits. Biochim Biophy Acta 1757:1019–1034
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125:1–15
Feng P, Zhao HB, Lu X (2015) Evolution of mitochondrial DNA and its relation to basal metabolic rate. Mitochondrial Dna 26:566–571
Fisher DO, Owens IP (2004) The comparative method in conservation biology. Trends Ecol Evol 19:391–398.
Galtier N, Enard D, Radondy Y, Bazin E, Belkhir K (2006) Mutation hot spots in mammalian mitochondrial DNA. Genome Res 16:215–222
Galtier N, Blier PU, Nabholz B (2009) Inverse relationship between longevity and evolutionary rate of mitochondrial proteins in mammals and birds. Mitochondrion 9:51–57
Garland T Jr, Harvey PH, Ives AR (1992) Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst Biol 41:18–32
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Harvey D, Pagel MD (1991) The Comparative Method in Evolutionary Biology. Oxford University Press, Oxford
Hasegawa M, Cao Y, Yang ZH (1998) Preponderance of slightly deleterious polymorphism in mitochondrial DNA: non-synonymous/synonymous rate ratio is much higher within species than between species. Mol Biol Evol 15:1499–1505
Hutter B, Bieg M, Helms V, Paulsen M (2010) Divergence of imprinted genes during mammalian evolution. BMC Evol Biol 10:1–10
Kujoth GC, Bradshaw P, Haroon S, Prolla T (2007) The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet 3:161–173
Li WH, Tanimura M, Sharp P (1987) An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol 25:330–342
Maddison WP, Maddison DRV (2009) Mesquite: a modular system for evolutionary analysis. Available at: http://mesquiteproject.org (Accessed 3 October 2011)
Martin AP ( 1995) Metabolic rate and directional nucleotide substitution in animal mitochondrial DNA. Mol Biol Evol 12:1124–1131
Martin AP, Palumbi S (1993) Body size, metabolic-rate, generation time, and the molecular clock. Proc Natl Acad Sci USA 90:4087–4091
McNab B (2008) An analysis of the factors that influence the level and scaling of mammalian BMR. Comp Biochem Physiol A Mol Integr Physiol 151:5–28
Mooers AO, Harvey P (1994) Metabolic rate, generation time, and the rate of molecular evolution in birds. Mol Phylogenet Evol 3:344–350
Müller DWH, Codron D, Werner J, Fritz J, Hummel J, Griebeler EM, Clauss M (2012) Dichotomy of eutherian reproduction and metabolism. Oikos 121:102–115
Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W (2007) Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res 17:413–421
Nabholz B, Glémin S, Galtier N (2008) Strong variations of mitochondrial mutation rate across mammals--the longevity hypothesis. Mol Biol Evol 25:120–130
Nunn GB, Stanley S (1998) Body size effects and rates of cytochrome b evolution in tube-nosed seabirds. Mol Biol Evol 15:1360–1371
Ohta T (1993) An examination of the generation-time effect on molecular evolution. Proc Natl Acad Sci USA 90:10676–10680
Rottenberg H (2006) Longevity and the evolution of the mitochondrial DNA-coded proteins in mammals. Mech Ageing Dev 127:748–760
Rottenberg H (2007a) Coevolution of exceptional longevity, exceptionally high metabolic rates, and mitochondrial DNA-coded proteins in mammals. Exp Gerontol 42:364–373
Rottenberg H (2007b) Exceptional longevity in songbirds is associated with high rates of evolution of cytochrome b, suggesting selection for reduced generation of free radicals. J Exp Biol 210:2170–2180
Rottenberg H (2009) Longevity and the early evolution of mtDNA-coded proteins in placental mammals. Mitochondrion 9:282
Rottenberg H (2014) Exceptional longevity and exceptionally high metabolic rates in anthropoid primates are linked to a major modification of the ubiquinone reduction site of cytochrome b. J Bioenerg Biomembr 46:435–445
Shen YY, Shi P, Sun YB, Zhang YP (2009) Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res 19:1760–1765
Speakman J (2005) Body size, energy metabolism and lifespan. J Exp Biol 208:1717–1730
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tranah GJ (2011) Mitochondrial-nuclear epistasis: implications for human aging and longevity. Ageing Res Rev 10:238–252
Wang Z, Yonezawa T, Liu B, Ma T, Shen X, Su J, Guo S, Hasegawa M, Liu J (2011) Domestication relaxed selective constraints on the yak mitochondrial genome. Mol Biol Evol 28:1553–1556
Yang ZH (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Acknowledgments
We thank two anonymous reviewers for their valuable and constructive comments. This research was supported by the National Natural Science Foundation of China (NSFC) (grant no. 31500310 to P.F.); the Scientific Research Foundation of the Higher Education Institutions of Guangxi Province, China (grant no. KY2015ZD016 to P.F.); a Start-up Fund, and a Development Support Program for Young Scholar from Guangxi Normal University to P.F.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
ESM 1
(DOCX 44 kb)
Rights and permissions
About this article
Cite this article
Feng, P., Zhou, Q. Absence of Relationship between Mitochondrial DNA Evolutionary Rate and Longevity in Mammals except for CYTB. J Mammal Evol 26, 1–7 (2019). https://doi.org/10.1007/s10914-017-9399-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10914-017-9399-4