Abstract
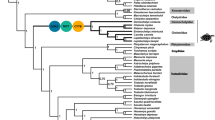
The superorder Cetartiodactyla has expanded to occupy a wide range of terrestrial and marine niches with very different energy requirements. As mitochondria are the primary site of energy production, we hypothesized that the structure and evolutionary rate of Cetartiodactyla mitochondrial DNA (mtDNA) have been influenced by the metabolic requirements of different ecological niches. In this study, we constructed full-length mtDNA sequences for four Cetartiodactyla species: Babyrousa babyrussa, Stenella clymene, Stenella frontalis, and Cephalorhynchus commersonii. We performed comparative mitochondrial genome analysis, selection pressure analyses, and phylogenetic independent contrasts (PIC) analysis to detect the structure and evolutionary rate of mitogenomes from Cetartiodactyla native to plain, plateau, and marine niches. We found that the structure of Cetartiodactyla mtDNA was consistent across three niches. However, selection pressure analyses showed that the evolutionary rate of mtDNA protein-coding genes (PCGs) was significantly different between niches. The marine Cetartiodactyla had the highest rate of PCG evolution, while in terrestrial species, the evolutionary rate of all PCGs except ND3 and ND6 was faster in plateau Cetartiodactyla than in plain Cetartiodactyla. PIC analysis also demonstrated that PCG evolutionary rates were positively correlated with niche type (R2 = 0.1226, p = 0.04239). This study suggests divergent mechanisms for the evolution of mtDNA PCGs, prompting the adaptation of Cetartiodactyla to diverse niches.








Similar content being viewed by others
Data availability
The data presented in this study are available in the article and supplementary material.
References
Alberdi, A., Gilbert, M. T. P., Razgour, O., Aizpurua, O., Aihartza, J., & Garin, I. (2015). Contrasting population-level responses to Pleistocene climatic oscillations in an alpine bat revealed by complete mitochondrial genomes and evolutionary history inference. Journal of Biogeography, 42(9), 1689–1700. https://doi.org/10.1111/jbi.12535
Allen, J. F. (2003). Cyclic, pseudocyclic and noncyclic photophosphorylation: New links in the chain. Trends in Plant Science, 8(1), 15–19. https://doi.org/10.1016/s1360-1385(02)00006-7
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., Pütz, J., Middendorf, M., & Stadler, P. F. (2013). MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69(2), 313–319. https://doi.org/10.1016/j.ympev.2012.08.023
Chang, H., Qiu, Z., Yuan, H., Wang, X., Li, X., Sun, H., Guo, X., Lu, Y., Feng, X., & Majid, M. (2020). Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Molecular Phylogenetics and Evolution, 145, 106734. https://doi.org/10.1016/j.ympev.2020.106734
Chi, Y., Wang, J., Xi, C., Qian, T., & Sheng, C. (2020). Spatial pattern of species richness among terrestrial mammals in China. Diversity, 12(3), 96. https://doi.org/10.3390/d12030096
Derous, D., Sahu, J., Douglas, A., Lusseau, D., & Wenzel, M. (2021). Comparative genomics of cetartiodactyla: Energy metabolism underpins the transition to an aquatic lifestyle. Conservation Physiology, 9(1), coaa136. https://doi.org/10.1093/conphys/coaa136
Dierckxsens, N., Mardulyn, P., & Smits, G. (2017). NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45(4). https://doi.org/10.1093/nar/gkw955
Drummond, A. J., Suchard, M. A., Xie, D., & Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29(8), 1969–1973. https://doi.org/10.1093/molbev/mss075
Düvel, K., Yecies, J. L., Menon, S., Raman, P., Lipovsky, A. I., Souza, A. L., Triantafellow, E., Ma, Q., Gorski, R., & Cleaver, S. (2010). Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell, 39(2), 171–183. https://doi.org/10.1016/j.molcel.2010.06.022.
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797. https://doi.org/10.1093/nar/gkh340
Felsenstein, & Joseph. (1985). Phylogenies and the comparative method. American Naturalist, 125(1), 1–15.
Foote, A. D., Morin, P. A., Pitman, R. L., Ávila-Arcos, M. C., Durban, J. W., van Helden, A., Sinding, M.-H.S., & Gilbert, M. T. P. (2013). Mitogenomic insights into a recently described and rarely observed killer whale morphotype. Polar Biology, 36(10), 1519–1523. https://doi.org/10.1007/s00300-013-1354-0
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., & Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Research, 32(suppl_2), W273–W279. https://doi.org/10.1093/nar/gkh458
Ghassemi-Khademi, T., & Hamidi, K. (2019). Re-evaluation of molecular phylogeny of the subfamily Cephalophinae (Bovidae: Artiodactyla); with notes on diversification of body size. Jordan Journal of Biological Sciences, 12(5).
Hassanin, A., Delsuc, F., Ropiquet, A., Hammer, C., van Vuuren, B. J., Matthee, C., Ruiz-Garcia, M., Catzeflis, F., Areskoug, V., & Nguyen, T. T. (2012). Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. Comptes Rendus Biologies, 335(1), 32–50. https://doi.org/10.1016/j.crvi.2011.11.002
Hassanin, A., Ropiquet, A., Couloux, A., & Cruaud, C. (2009). Evolution of the mitochondrial genome in mammals living at high altitude: New insights from a study of the tribe Caprini (Bovidae, Antilopinae). Journal of Molecular Evolution, 68(4), 293–310. https://doi.org/10.1007/s00239-009-9208-7
Humphreys, A. M., & Barraclough, T. G. (2014). The evolutionary reality of higher taxa in mammals. Proceedings of the Royal Society b: Biological Sciences, 281(1783), 20132750. https://doi.org/10.1098/rspb.2013.2750
Hüttemann, M., Lee, I., Samavati, L., Yu, H., & Doan, J. W. (2007). Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1773(12), 1701–1720. https://doi.org/10.1016/j.bbamcr.2007.10.001
Iommarini, L., Ghelli, A., Gasparre, G., & Porcelli, A. M. (2017). Mitochondrial metabolism and energy sensing in tumor progression. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 1858(8), 582–590. https://doi.org/10.1016/j.bbabio.2017.02.006
Jessop, N., & Herrero, M. (1998). Modelling fermentation in an in vitro gas production system: Effects of microbial activity. BSAP Occasional Publication, 22, 81–84. https://doi.org/10.1017/S0263967X00032304
Koch, R. E., Buchanan, K. L., Casagrande, S., Crino, O., Dowling, D. K., Hill, G. E., Hood, W. R., McKenzie, M., Mariette, M. M., & Noble, D. W. (2021). Integrating mitochondrial aerobic metabolism into ecology and evolution. Trends in Ecology & Evolution. https://doi.org/10.1016/j.tree.2020.12.006
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547. https://doi.org/10.1093/molbev/msy096
Letunic, I., & Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Research, 47(W1), W256–W259. https://doi.org/10.1093/nar/gkz239
Li, F., Lv, Y., Wen, Z., Bian, C., Zhang, X., Guo, S., Shi, Q., & Li, D. (2021). The complete mitochondrial genome of the intertidal spider (Desis jiaxiangi) provides novel insights into the adaptive evolution of the mitogenome and the evolution of spiders. BMC Ecology and Evolution, 21(1), 1–15. https://doi.org/10.1186/s12862-021-01803-y
Li, G.-G., Zhang, M.-X., Swa, K., Maung, K.-W., & Quan, R.-C. (2017). Complete mitochondrial genome of the leaf muntjac (Muntiacus putaoensis) and phylogenetics of the genus Muntiacus. Zoological Research, 38(5), 310. https://doi.org/10.24272/j.issn.2095-8137.2017.058
Maes, D., Collins, D., Declercq, L., Foyouzi-Yousseffi, R., Gan, D., Mammone, T., Pelle, E., Marenus, K., & Gedeon, H. (2004). Improving cellular function through modulation of energy metabolism. International Journal of Cosmetic Ence, 26(5), 268–269. https://doi.org/10.1111/j.1467-2494.2004.00230_4.x
Mendes, S., Newton, J., Reid, R. J., Zuur, A. F., & Pierce, G. J. (2007). Stable carbon and nitrogen isotope ratio profiling of sperm whale teeth reveals ontogenetic movements and trophic ecology. Oecologia, 151(4), 605–615. https://doi.org/10.1007/s00442-006-0612-z
Monnens, M., Thijs, S., Briscoe, A. G., Clark, M., Frost, E. J., Littlewood, D. T. J., Sewell, M., Smeets, K., Artois, T., & Vanhove, M. P. (2020). The first mitochondrial genomes of endosymbiotic rhabdocoels illustrate evolutionary relaxation of atp8 and genome plasticity in flatworms. International Journal of Biological Macromolecules, 162, 454–469. https://doi.org/10.1016/j.ijbiomac.2020.06.025
Mori, S., & Matsunami, M. (2018). Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes & Genetic Systems. https://doi.org/10.1266/ggs.17-00015
Nishida, A. H., & Ochman, H. (2018). Rates of gut microbiome divergence in mammals. Molecular Ecology, 27(8), 1884–1897. https://doi.org/10.1111/mec.14473
Olsen, C., Qaadri, K., Moir, R., Kearse, M., Buxton, S., & Cheung, M. (2014). Geneious R7: A bioinformatics platform for biologists. International plant and animal genome conference Xxii,
Park, H., Lee, J.-Y., Lim, W., & Song, G. (2021). Assessment of the in vivo genotoxicity of pendimethalin via mitochondrial bioenergetics and transcriptional profiles during embryogenesis in zebrafish: Implication of electron transport chain activity and developmental defects. Journal of Hazardous Materials, 411, 125153. https://doi.org/10.1016/j.jhazmat.2021.125153
Perna, N. T., & Kocher, T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. Journal of Molecular Evolution, 41(3), 353–358. https://doi.org/10.1007/BF00186547
Priyono, D. S., Solihin, D. D., Farajallah, A., & Purwantara, B. (2020). The first complete mitochondrial genome sequence of the endangered mountain anoa (Bubalus quarlesi)(Artiodactyla: Bovidae) and phylogenetic analysis. Journal of Asia-Pacific Biodiversity, 13(2), 123–133. https://doi.org/10.1016/j.japb.2020.01.006
Qiu, Q., Zhang, G., Ma, T., Qian, W., Wang, J., Ye, Z., Cao, C., Hu, Q., Kim, J., & Larkin, D. M. (2012). The yak genome and adaptation to life at high altitude. Nature Genetics, 44(8), 946–949. https://doi.org/10.1038/ng.2343
Ramos, E. K., & d. S., Freitas, L., & Nery, M. F. (2020). The role of selection in the evolution of marine turtles mitogenomes. Scientific Reports, 10(1), 1–13. https://doi.org/10.1038/s41598-020-73874-8
Ritchie, A. M., Lo, N., & Ho, S. Y. (2016). Examining the sensitivity of molecular species delimitations to the choice of mitochondrial marker. Organisms Diversity & Evolution, 16(3), 467–480. https://doi.org/10.1007/s13127-016-0275-5
Sanno, R., Kataoka, K., Hayakawa, S., Ide, K., Nguyen, C. N., Nguyen, T. P., Le, B. T., Kim, O. T., Mineta, K., & Takeyama, H. (2021). Comparative analysis of mitochondrial genomes in Gryllidea (Insecta: Orthoptera): Implications for adaptive evolution in ant-loving crickets. Genome Biology and Evolution, 13(10), evab222. https://doi.org/10.1093/gbe/evab222
Sarvani, R. K., Parmar, D. R., Tabasum, W., Thota, N., Sreenivas, A., & Gaur, A. (2018). Characterization of the complete mitogenome of Indian Mouse Deer, Moschiola indica (Artiodactyla: Tragulidae) and its evolutionary significance. Science and Reports, 8(1), 1–12. https://doi.org/10.1038/s41598-018-20946-5
Shen, T., Xu, S., Wang, X., Yu, W., Zhou, K., & Yang, G. (2012). Adaptive evolution and functional constraint at TLR4 during the secondary aquatic adaptation and diversification of cetaceans. BMC Evolutionary Biology, 12(1), 1–12. https://doi.org/10.1186/1471-2148-12-39
Shen, Y.-Y., Shi, P., Sun, Y.-B., & Zhang, Y.-P. (2009). Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Research, 19(10), 1760–1765. https://doi.org/10.1101/gr.093138.109
Shin, S., & Chung, E. (2016). Identification of differentially expressed genes between high and low marbling score grades of the longissimus lumborum muscle in Hanwoo (Korean cattle). Meat Science, 121, 114–118. https://doi.org/10.1016/j.meatsci.2016.05.018
Soares, P., Abrantes, D., Rito, T., Thomson, N., Radivojac, P., Li, B., Macaulay, V., Samuels, D. C., & Pereira, L. (2013). Evaluating purifying selection in the mitochondrial DNA of various mammalian species. PLoS ONE, 8(3), e58993. https://doi.org/10.1371/journal.pone.0058993
Sun, J. T., Jin, P. Y., Hoffmann, A., Duan, X. Z., Dai, J., Hu, G., Xue, X. F., & Hong, X. Y. (2018). Evolutionary divergence of mitochondrial genomes in two Tetranychus species distributed across different climates. Insect Molecular Biology, 27(6), 698–709. https://doi.org/10.1111/imb.12501
Tan, Z.-D., Shen, L.-Y., Cheng, X., Gan, M.-L., Zhang, S.-H., & Zhu, L. (2018). The complete mitochondrial genome sequence of Changbai Mountains wild boar (Cetartiodactyla: Suidae). Conservation Genetics Resources, 10(1), 99–102. https://doi.org/10.1007/s12686-017-0773-6
Tomasco, I. H., & Lessa, E. P. (2011). The evolution of mitochondrial genomes in subterranean caviomorph rodents: Adaptation against a background of purifying selection. Molecular Phylogenetics and Evolution, 61(1), 64–70. https://doi.org/10.1016/j.ympev.2011.06.014
Wallace, D. C. (2007). Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annual Review of Biochemistry, 76, 781–821. https://doi.org/10.1146/annurev.biochem.76.081205.150955
Wang, X., Chen, Y., Shang, Y., Wu, X., Wei, Q., Chen, J., Yan, J., & Zhang, H. (2019). Comparison of the gut microbiome in red deer (Cervus elaphus) and fallow deer (Dama dama) by high-throughput sequencing of the V3–V4 region of the 16S rRNA gene. ScienceAsia, 45(6), 515–524. https://doi.org/10.2306/scienceasia1513-1874.2019.45.515
Wang, X., Zhou, S., Wu, X., Wei, Q., Shang, Y., Sun, G., Mei, X., Dong, Y., Sha, W., & Zhang, H. (2021). High-altitude adaptation in vertebrates as revealed by mitochondrial genome analyses. Ecology and Evolution, 11(21), 15077–15084. https://doi.org/10.1002/ece3.8189
Wang, Y., Shen, Y., Feng, C., Zhao, K., Song, Z., Zhang, Y., Yang, L., & He, S. (2016). Mitogenomic perspectives on the origin of Tibetan loaches and their adaptation to high altitude. Science and Reports, 6(1), 1–10. https://doi.org/10.1038/srep29690
Wei, Q., Dong, Y., Sun, G., Wang, X., Wu, X., Gao, X., Sha, W., Yang, G., & Zhang, H. (2021). FGF gene family characterization provides insights into its adaptive evolution in Carnivora. Ecology and Evolution, 11(14), 9837–9847. https://doi.org/10.1002/ece3.7814
Wei, Q., Zhang, H., Wu, X., & Sha, W. (2020). The selective constraints of ecological specialization in mustelidae on mitochondrial genomes. Mammal Research, 65(1), 85–92. https://doi.org/10.1007/s13364-019-00461-2
Wu, X., Wei, Q., Wang, X., Shang, Y., & Zhang, H. (2022). Evolutionary and dietary relationships of wild mammals based on the gut microbiome. Gene, 808, 145999. https://doi.org/10.1016/j.gene.2021.145999
Yang, M., Gong, L., Sui, J., & Li, X. (2019). The complete mitochondrial genome of Calyptogena marissinica (Heterodonta: Veneroida: Vesicomyidae): Insight into the deep-sea adaptive evolution of vesicomyids. PLoS ONE, 14(9), e0217952. https://doi.org/10.1371/journal.pone.0217952
Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24(8), 1586–1591. https://doi.org/10.1093/molbev/msm088
Yuan, Y., Zhang, Y., Zhang, P., Liu, C., Wang, J., Gao, H., Hoelzel, A. R., Seim, I., Lv, M., & Lin, M. (2021). Comparative genomics provides insights into the aquatic adaptations of mammals. Proceedings of the National Academy of Sciences, 118(37). https://doi.org/10.1073/pnas.2106080118
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., & Wang, G. T. (2020). PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20(1), 348–355. https://doi.org/10.1111/1755-0998.13096
Zhang, Z., Mu, Y., Shan, L., Sun, D., Guo, W., Yu, Z., Tian, R., Xu, S., & Yang, G. (2019). Divergent evolution of TRC genes in mammalian niche adaptation. Frontiers in Immunology, 10, 871. https://doi.org/10.3389/fimmu.2019.00871
Zhang, Z., Xu, D., Wang, L., Hao, J., Wang, J., Zhou, X., Wang, W., Qiu, Q., Huang, X., Zhou, J., Long, R., Zhao, F., & Shi, P. (2016, Jul 25). Convergent evolution of rumen microbiomes in high-altitude mammals. Current Biology, 26(14), 1873–1879. https://doi.org/10.1016/j.cub.2016.05.012
Funding
This work was supported by the National Natural Science Foundation of China (31872242, 32070405, 32001228, 32000291, 32170530).
Author information
Authors and Affiliations
Contributions
Conceptualization, H. Z., X. Wang, X. Wu, and Q. W.; methodology, X. Wang, Y. S., S. Z., and G. S.; writing—original draft preparation, X. Wang; writing—reviewing and editing, X. Wang; formal analysis, X. Wang, S. M., H. D., and W. S.; validation, X. Wang; funding acquisition, H. Z., X. Wu, and W. S. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Shang, Y., Wu, X. et al. Divergent evolution of mitogenomics in Cetartiodactyla niche adaptation. Org Divers Evol 23, 243–259 (2023). https://doi.org/10.1007/s13127-022-00574-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-022-00574-8