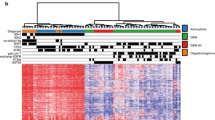
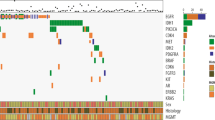
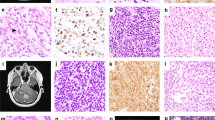
Abstract
A prompt and reliable molecular diagnosis for brain tumors has become crucial in precision medicine. While Comprehensive Genomic Profiling (CGP) has become feasible, there remains room for enhancement in brain tumor diagnosis due to the partial lack of essential genes and limitations in broad copy number analysis. In addition, the long turnaround time of commercially available CGPs poses an additional obstacle to the timely implementation of results in clinics. To address these challenges, we developed a CGP encompassing 113 genes, genome-wide copy number changes, and MGMT promoter methylation. Our CGP incorporates not only diagnostic genes but also supplementary genes valuable for research. Our CGP enables us to simultaneous identification of mutations, gene fusions, focal and broad copy number alterations, and MGMT promoter methylation status, with results delivered within a minimum of 4 days. Validation of our CGP, through comparisons with whole-genome sequencing, RNA sequencing, and pyrosequencing, has certified its accuracy and reliability. We applied our CGP for 23 consecutive cases of intracranial mass lesions, which demonstrated its efficacy in aiding diagnosis and prognostication. Our CGP offers a comprehensive and rapid molecular profiling for gliomas, which could potentially apply to clinical practices and research primarily in the field of brain tumors.





Similar content being viewed by others
Data availability
Raw sequencing data of this study are available from the corresponding author on request.
References
Aoyagi Y, Kano Y, Tohyama K et al (2022) Clinical utility of comprehensive genomic profiling in Japan: result of PROFILE-F study. PLoS ONE 17(3):e0266112
Appay R, Dehais C, Maurage CA et al (2019) CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol 21(12):1519–1528
Bady P, Sciuscio D, Diserens A-C et al (2012) MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol 124(4):547–560
Ballester LY, Fuller GN, Powell SZ et al (2017) Retrospective analysis of molecular and immunohistochemical characterization of 381 primary brain tumors. J Neuropathol Exp Neurol 76(3):179–188
Bell EH, Zhang P, Shaw EG et al (2020) Comprehensive genomic analysis in NRG Oncology/RTOG 9802: a Phase III trial of radiation versus radiation plus procarbazine, lomustine (CCNU), and vincristine in high-risk low-grade glioma. J Clin Oncol 38(29):3407–3417
Beroukhim R, Mermel CH, Porter D et al (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463(7283):899–905
Brennan CW, Verhaak RG, McKenna A et al (2013) The somatic genomic landscape of glioblastoma. Cell 155(2):462–477
Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG et al (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372(26):2481–2498
Christians A, Hartmann C, Benner A et al (2012) Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoS ONE 7(3):e33449
Cibulskis K, Lawrence MS, Carter SL et al (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31(3):213–219
Clarke M, Mackay A, Ismer B et al (2020) Infant high-grade gliomas comprise multiple subgroups characterized by novel targetable gene fusions and favorable outcomes. Cancer Discov 10(7):942–963
DeSisto J, Lucas JT Jr, Xu K et al (2021) Comprehensive molecular characterization of pediatric radiation-induced high-grade glioma. Nat Commun 12(1):5531
Do H, Dobrovic A (2015) Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem 61(1):64–71
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21
Ellison DW, Aldape KD, Capper D et al (2020) cIMPACT-NOW update 7: advancing the molecular classification of ependymal tumors. Brain Pathol 30(5):863–866
Esteller M, Hamilton SR, Burger PC et al (1999) Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 59(4):793–797
Frattini V, Trifonov V, Chan JM et al (2013) The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 45(10):1141–1149
Guerreiro Stucklin AS, Ryall S, Fukuoka K et al (2019) Alterations in ALK/ROS1/NTRK/MET drive a group of infantile hemispheric gliomas. Nat Commun 10(1):4343
Guo Q, Lakatos E, Bakir IA et al (2022) The mutational signatures of formalin fixation on the human genome. Nat Commun 13(1):4487
Hatanaka Y, Kuwata T, Morii E et al (2021) The Japanese Society of Pathology Practical Guidelines on the handling of pathological tissue samples for cancer genomic medicine. Pathol Int 71(11):725–740
Hegi ME, Liu L, Herman JG et al (2008) Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol 26(25):4189–4199
Higa N, Akahane T, Yokoyama S et al (2020) A tailored next-generation sequencing panel identified distinct subtypes of wildtype IDH and TERT promoter glioblastomas. Cancer Sci 111(10):3902–3911
Ji MS, Eldred BSC, Liu R et al (2020) Targeted next-generation sequencing of 565 neuro-oncology patients at UCLA: a single-institution experience. Neurooncol Adv 2(1):vdaa009
Kohno T, Kato M, Kohsaka S et al (2022) C-CAT: the national datacenter for cancer genomic medicine in Japan. Cancer Discov 12(11):2509–2515
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27(11):1571–1572
Li BK, Vasiljevic A, Dufour C et al (2020) Pineoblastoma segregates into molecular sub-groups with distinct clinico-pathologic features: a Rare Brain Tumor Consortium registry study. Acta Neuropathol 139(2):223–241
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26(5):589–595
Lopez GY, Van Ziffle J, Onodera C et al (2019) The genetic landscape of gliomas arising after therapeutic radiation. Acta Neuropathol 137(1):139–150
Lorenz J, Rothhammer-Hampl T, Zoubaa S et al (2020) A comprehensive DNA panel next generation sequencing approach supporting diagnostics and therapy prediction in neurooncology. Acta Neuropathol Commun 8(1):124
Louis DN, Perry A, Wesseling P et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1):10–12
Malley DS, Hamoudi RA, Kocialkowski S et al (2011) A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol 121(5):651–661
Nassar LR, Barber GP, Benet-Pages A et al (2023) The UCSC genome browser database: 2023 update. Nucleic Acids Res 51(D1):D1188–D1195
Nikiforova MN, Wald AI, Melan MA et al (2016) Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors. Neuro Oncol 18(3):379–387
Ostrom QT, Gittleman H, Fulop J et al (2015) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol 17(Suppl 4):iv1–iv62
Palmisano WA, Divine KK, Saccomanno G et al (2000) Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 60(21):5954–5958
Paugh BS, Qu C, Jones C et al (2010) Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol 28(18):3061–3068
Pekmezci M, Villanueva-Meyer JE, Goode B et al (2018) The genetic landscape of ganglioglioma. Acta Neuropathol Commun 6(1):47
Phi JH, Park AK, Lee S et al (2018) Genomic analysis reveals secondary glioblastoma after radiotherapy in a subset of recurrent medulloblastomas. Acta Neuropathol 135(6):939–953
Rausch T, Zichner T, Schlattl A et al (2012) DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28(18):i333–i339
Roosen M, Odé Z, Bunt J, Kool M (2022) The oncogenic fusion landscape in pediatric CNS neoplasms. Acta Neuropathol 143(4):427–451
Sahm F, Schrimpf D, Jones DT et al (2016) Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol 131(6):903–910
Sanson M, Marie Y, Paris S et al (2009) Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 27(25):4150–4154
Saunders CT, Wong WS, Swamy S et al (2012) Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28(14):1811–1817
Shirahata M, Ono T, Stichel D et al (2018) Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol 136(1):153–166
Shiraishi Y, Sato Y, Chiba K et al (2013) An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res 41(7):e89
Siddaway R, Milos S, Vadivel AKA et al (2022) Splicing is an alternate oncogenic pathway activation mechanism in glioma. Nat Commun 13(1):588
Stupp R, Hegi ME, Mason WP et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10(5):459–466
Sturm D, Orr BA, Toprak UH et al (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164(5):1060–1072
Suzuki H, Aoki K, Chiba K et al (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47(5):458–468
Synhaeve NE, van den Bent MJ, French PJ et al (2018) Clinical evaluation of a dedicated next generation sequencing panel for routine glioma diagnostics. Acta Neuropathol Commun 6(1):126
Tabone T, Abuhusain HJ, Nowak AK et al (2014) Multigene profiling to identify alternative treatment options for glioblastoma: a pilot study. J Clin Pathol 67(7):550–555
Taylor AM, Shih J, Ha G et al (2018) Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33(4):676–689.e3
Tomlins SA, Hovelson DH, Suga JM et al (2021) Real-world performance of a comprehensive genomic profiling test optimized for small tumor samples. JCO Precis Oncol 5
Uhrig S, Ellermann J, Walther T et al (2021) Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res 31(3):448–460
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164
Wang Y, Zhao Y, Bollas A et al (2021) Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol 39(11):1348–1365
Wefers AK, Stichel D, Schrimpf D et al (2020) Isomorphic diffuse glioma is a morphologically and molecularly distinct tumour entity with recurrent gene fusions of MYBL1 or MYB and a benign disease course. Acta Neuropathol 139(1):193–209
Wick W, Gorlia T, Bendszus M et al (2017) Lomustine and bevacizumab in progressive glioblastoma. N Engl J Med 377(20):1954–1963
Yang RR, Shi ZF, Zhang ZY et al (2020) IDH mutant lower grade (WHO Grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol 30(3):541–553
Acknowledgements
HS is supported by the Japan Society for the Promotion of Science KAKENHI (22H03190, 22K19591), National Cancer Center Research and Development Funds (2021-A-1), Japan Agency for Medical Research and Development (22ck0106693h0002, 23ck0106877h0001), MSD Life Science Foundation, Public Interest Incorporated Foundation, Japan Science and Technology Agency (21467119), Takeda Science Foundation, the Uehara Memorial Foundation, the Cell Science Research Foundation, and Astellas Foundation for Research on Metabolic Disorders. A.U. is supported by Kurozumi Medical Foundation. We thank National Cancer Center Research Institute Core Facility and Mai Honda-Kitahara for performing a part of panel-based sequencing. Blood samples of NCCH cases were provided by the National Cancer Center Biobank, Japan. Both the Core Facility and Biobank were supported in part by National Cancer Center Research and Development Fund, Japan. The super-computing resource was provided by Human Genome Center, The Institute of Medical Science, The University of Tokyo, and the NIG supercomputer at ROIS National Institute of Genetics.
Author information
Authors and Affiliations
Contributions
HS led the study. TN and YF performed the bioinformatics analyses. TN, YF, SN, AU, RY, and HS designed a gene panel. MO, MT, TU, YA, RH, JI, KY, RS, and YN provided the patient material and expert advice. TN, RY, and HS prepared the manuscript, figures, and tables. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest with this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nakashima, T., Yamamoto, R., Ohno, M. et al. Development of a rapid and comprehensive genomic profiling test supporting diagnosis and research for gliomas. Brain Tumor Pathol 41, 50–60 (2024). https://doi.org/10.1007/s10014-023-00476-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10014-023-00476-3