Abstract
T helper 2 (Th2) cytokine production by invariant natural killer T (iNKT) cells is involved in the development of asthma, but the regulation of Th2 cytokines in iNKT cells remains unknown. Although it is known that progranulin (PGRN) induces the production of Th2 cytokines in iNKT cells in vivo, the underlying mechanism is not clear. This study aims to investigate the role of PGRN in iNKT cells. The effects of PGRN on the differentiation of iNKT cells was detected by flow cytometry. Then stimulation of iNKT cells and airway resistance were carried out to evaluate the function of PGRN on iNKT cells. Furthermore, the mechanisms of PGRN in regulating iNKT cells was investigated by RT-PCR, WB, confocal and luciferase reporter assays. The absolute number of iNKT cells decreased in PGRN KO mice despite an increase in the percentage of iNKT cells. Furthermore, analyzing the subsets of iNKT cells, we found that NKT2 cells and their IL-4 production were reduced. Mechanistically, the decrease in NKT2 cells in the PGRN KO mice was caused by increased expression of enhancer of zeste homolog 2 (EZH2), that in turn caused increased degradation and altered nuclear localization of PLZF. Interestingly, PGRN signaling decreased expression of EZH2 and treatment of the PGRN KO mice with the EZH2 specific inhibitor GSK343 rescued the defect in NKT2 differentiation, IL-4 generation, and PLZF expression. Altogether, We have revealed a new pathway (PGRN-EZH2-PLZF), which regulates the Th2 responses of iNKT cells and provides a potentially new target for asthma treatment.
Similar content being viewed by others
Introduction
Asthma is one of the most common chronic inflammatory lung diseases in children. The development of asthma involves many types of immune cells such as T cells, B cells, eosinophils, and neutrophils [1]. T helper 2 (Th2) cytokine production plays a major role in the pathogenesis of asthma. Th2 lymphocytes also increase the synthesis of IgE by stimulating humoral immune responses [2]. Invariant natural killer T (iNKT) cells are characterized by the expression of an invariant T-cell receptor (TCR) α chain consisting of Vα14/Jα18 in mice or Vα24/Jα18 in humans that recognizes glycolipid antigens, releasing large amounts of type 1 and type 2 cytokines. iNKT cells drive the development of asthma-like lung pathology in mice [3], and an increase in iNKT cells has been found in asthma patients after an allergen challenge [4], indicating that iNKT cells play an important role in asthma. Although the role of iNKT cells in asthma has been studied for many years, how iNKT cells regulate the pathogenesis of this disease has not been clearly elucidated.
Progranulin (PGRN), also known as granulin-epithelin precursor, was first recognized as a growth factor in wound healing. Subsequently, PGRN was found to be expressed in other cell types including macrophages, neurons, and chondrocytes [5]. In rheumatoid arthritis (RA), PGRN plays an anti-inflammatory role by binding to tumor necrosis factor (TNF) receptors, which antagonizes TNF-α signaling [6, 7]. Additionally, the level of serum PGRN is lower in patients with asthma than in healthy controls [8]. Furthermore, macrophage-derived PGRN induces NKT cell production of Th2 cytokines in mice, which is critical during the development of allergic airway inflammation [9]. However, how PGRN regulates iNKT cells and their production of Th2 cytokines is still unknown.
Enhancer of zeste homolog 2 (EZH2) belongs to the polycomb repressive complex 2 (PRC2) protein family and methylates lysine 27 of histone-H3 (H3K27me3) [10]. It also modifies non-histone proteins such as VAV1 and TALIN, which is important for actin polymerization and cell migration [11, 12]. Previous studies have indicated that EZH2 suppresses iNKT cell induction of Th2 cytokines that drive asthma-like lung pathology in mice, which occurs via methylation-dependent ubiquitination of promyelocytic leukemia zinc finger (PLZF) in iNKT cells [13]. Both PGRN and EZH2 have the ability to regulate iNKT cells and Th2 cytokine production, but their relationship has not been elucidated to date.
In this study, we investigated the molecular mechanism of the regulation of Th2 responses by PGRN in iNKT cells using PGRN knockout (KO) mice. In these mice, we found that the iNKT cell number and IL-4 production were decreased despite an increase in the percentage of iNKT cells. Interestingly, EZH2 expression was increased and led to downregulation of PLZF. Furthermore, expression of the EZH2 inhibitor GSK343 could rescue the defect in NKT2 cell differentiation, IL-4 production, and PLZF expression. All these studies have clarified a novel pathway whereby PGRN regulates NKT2 cells via upregulating EZH2 expression. This study also identifies a potentially new therapeutic target for asthma treatment.
Materials and methods
Mice
For experiments, 6-8-week-old PGRN−/− mice (PGRN KO) (Jackson Laboratory) and age-matched C57BL/6 wild-type (WT) mice (controls) were used. All mice were kept in individually ventilated cages at the Children’s Hospital of Chongqing Medical University. All animal work was reviewed and approved by the Institutional Animal Care and Use Committee of the Children’s Hospital of Chongqing Medical University and all experiments were performed according to the principles of medical ethics and the requirements of the Declaration of Helsinki.
Flow cytometry
Splenocytes and thymocytes were isolated based on a previously published protocol [14]. For staining surface markers, the cells were incubated with the following monoclonal antibodies in PBS + 2%FBS at room temperature for 30 min: anti-CD44 (clone: IM7), anti-Annexin V, anti-CD4 (clone: RM4-5), anti-NK1.1 (clone: PK136), anti-TCR-β (clone: H57-597), anti-CD24 (clone: M1/69), anti-CD8a (clone: 53-6.7), anti-CD45.2 (clone: 104), and anti-CD45.1 (clone: A20). All antibodies above were purchased from BioLegend.
For intracellular staining, the cells were fixed and permeabilized using a Foxp3 Staining Buffer Set (eBioscience) according to the manufacturer’s instructions and then stained with APC-anti-RORγt (clone: AFKJ5-9, eBioscience), anti-T-bet (clone: PK136, BioLegend), anti-GATA3 (clone: 10E10A23, BioLegend), anti-Ki-67 (clone: SolA15, eBioscience), anti-EZH2 (clone: D2C9, Cell Signaling Technology), anti-PGRN (clone:ERP18539-59, abcam), anti-PLZF (D-9) (mouse) (Santa Cruz), PE/Cyanine7 anti-mouse PLZF (clone: 9E12, BioLgend) and FITC-labeled rat anti-mouse IgG1 secondary antibody (clone: M1-14D12, eBioscience).
For detection of phosphorylated proteins, iNKT cells were labeled and pulsed with phorbol 12-myristate 13-acetate (PMA, 50 ng/ml) and ionomycin (500 ng/ml) at 37 °C for different times. After activation, the cells were fixed and permeabilized with Phosflow Perm Buffer III (BD Biosciences) and then stained with anti-phospho-NF-κB P65 (Ser536) rabbit mAb (clone: 93H1) and anti-phospho-IKKα/β (Ser176/180) (clone: 16A6) rabbit mAb. All primary antibodies were from Cell Signaling Technology. AF488-goat anti-rabbit antibody (Life Technologies) was used as the secondary antibody.
Bone marrow chimeric mice
For chimera studies, bone marrow cells (1 × 107) from WT or PGRN−/− mice and CD45.1 mice were mixed 1:1 and then intravenously injected into 6 weeks old female irradiated (6 Gy) CD45.1 mice recipients randomly. After 8 weeks, the recipient mice were euthanized for experiments.
Stimulation of iNKT cells
For in vitro stimulation with alpha-galactosylceramide (α-Galcer), thymocytes were isolated and seeded in 24-well plates in RPMI + 10% FBS with or without 125 ng/ml α-Galcer for 72 h, incubating with 50 ng/ml PMA and 500 ng/ml ionomycin for the last 5 h. The cells were then stained for surface markers using mCd1d-PBS57-tetramer, anti-7AAD (BBI Life Sciences), and anti-TCR-β (clone: H57-597, BioLegend) and stained for intracellular markers using anti-mouse TNF-α (clone: MP6-XT22, BioLegend), anti-IL-4 (clone: 11B11, BioLegend), anti-IFN-γ (clone: XMG1.2, BioLegend), and anti-IL-17A (clone: TC11-18H10.1, BioLegend). For anti-PGRN stimulation, iNKT cells were isolated from WT mice, stimulated with or without anti-PGRN antibodies, and then stained for EZH2. The expression levels of EZH2 were analyzed by flow cytometry.
Immunoblotting
For detection of PLZF and EZH2 expression in thymus CD4+ cells, the cells (3 × 106) were sorted from the thymus glands of WT and PGRN KO mice. After lysis and protein denaturation, the cell extracts were subjected to SDS-PAGE and then immunoblotted using an anti-PLZF (Cell Signaling Technology, Cat: 39784) and anti-EZH2 (Cell Signaling Technology, Cat: 5246) antibody. β-Actin was used as the loading control.
Plasmid construction
The promoters of mEzh2 and mPlzf were inserted into the pGL3-Basic vector named pGL3-mEZH2 promoter-luciferase and pGL3-mPLZF promoter-luciferase, these PCR primers were as follows: mEzh2 promoter primer forward: 5ʹ-CATTTAGTATAGCTAGACAGAAATGT-3ʹ, mEzh2 promoter primer reverse: 5ʹ-GGATTATTCTAAAAGCAATGATT-3ʹ, mEzh2 promoter primer forward: 5ʹ-GTACCATTTAGTATAGCTAGACAGAAAT-3ʹ, mEzh2 promoter primer reverse: 5ʹ-TCGAGGATTATTCTAAAAGCAATGAT-3ʹ, mPlzf promoter primer forward: 5ʹ-CTCCCAAGTTTGCCAAAGTCC-3ʹ. mPlzf promoter primer reverse: 5ʹ-GGATGCTCCCCTGGGCT-3ʹ, mPlzf promoter primer forward: 5ʹ-GTACCTCCCAAGTTTGCCAAA-3ʹ, mPlzf promoter primer reverse: 5ʹ-TCGAGGATGCTCCCCTGG-3ʹ. The gene of Ezh2 was inserted into the pAd5-E1-CMV-Flag named pAd5-E1-CMV-mEZH2-flag, the primer sequences were as follows: mEzh2 primer forward: 5ʹ-ATATGGGCCAGACTGGGAA-3ʹ, mEzh2 primer reverse: 5ʹ-CTAGTAGGGATTTCCATTTCTCGT-3ʹ, mEzh2 primer forward: 5ʹ-CGATATGGGCCAGACTGGIGAAGAA-3ʹ, mEzh2 primer reverse: 5ʹ-TAGGGATTTCCATTTCTCGTTCG-3ʹ. The gene of mPgrn was inserted into the pAd5-E1-CMV named pAd5-E1-CMV-mPGRN, the primer sequences were as follows: mPgrn primer forward: 5ʹ-ATCGATATGTGGGTCCTGATGAGCTGGCTG-3ʹ. mPgrn primer reverse: 5ʹ-TACTAGTCAGTAGCGGTCTTGGGACCGGAT-3ʹ.
Luciferase reporter assay
To investigate whether PGRN regulates the promoter of Ezh2, HEK293T cells were cultured in 24-well plates and co-transfected with pGL3-mEZH2 promoter-luciferase and pAd5-E1-CMV or pAd5-E1-CMV-mPGRN. To investigate whether EZH2 or PGRN regulates the promoter of Plzf, HEK293T cells were cultured in 24-well plates and co-transfected with pGL3-mPLZF promoter-luciferase and pAd5-E1-CMV, pAd5-E1-CMV-mEZH2-Flag, or pAd5-E1-CMV-mPGRN-Flag, all groups were transfected simultaneously pCMV-RL and using transfection reagents lip3000 (Invitrogen). After 48 hours incubation, passive lysis buffer was added to each well and the supernatant was collected for detection in accordance with the Dual-Luciferase®Report Assay Systems Kit (Solarbio) [15].
Quantitative RT-PCR
Thymocytes were stained with mCd1d-PBS57-tetramer and anti-TCR-β and then the iNKT cells were sorted using a BD FACS Aria III Cell Sorter. Total RNA from the sorted iNKT cells was isolated using Trizol Reagent (BioTeke) and reverse transcribed into cDNA with a PrimeScript RT reagent Kit (US, Takara). Gene expression was then measured with a CFX96 Real-Time PCR System (Bio-Rad) using the following primer pairs:
Irf4: F 5ʹ-GGAAGACAAGATTACGATGTGC-3ʹ; R 5ʹ-AATCCTGTACACCTTGTATGGG-3ʹ
Egr2: F 5ʹ-GATCCTTCAGCATTCTTATCGC-3ʹ; R 5ʹ-GATCATAGGAATGAGACCTGGG-3ʹ
Ezh2: F 5ʹ-ATGAAGCAGACAGAAGAGGAAA-3ʹ; R 5ʹ-GGATAGCCCTCTTAGCAAAGAT-3ʹ
Zbtb16: F 5ʹ-CGCCACCTTCGCTCACATACAG-3ʹ; R 5ʹ-TGGTGCTTGAGGCTGAACTTCTTG-3ʹ.
Confocal microscopy
Confocal microscopy was performed as previously described [16]. Sorted iNKT cells were added to slides coated with poly-lysine. After fixation and permeabilization, the cells were stained with anti-PLZF (Santa Cruz Biotechnology), anti-EZH2 (clone: D2C9, Cell Signaling Technology), and AF488-phallodin (Invitrogen) for 1 h. AF546-goat anti-mouse antibody (1:400) was used as the secondary antibody. For staining cell nuclei, 1.5 μg/ml of DAPI (Beyotime) was used. For stimulation, the iNKT cells were incubated with α-Galcer (125 ng/ml) in 96-well plates for 72 h and then stained as described above. The images were obtained by confocal fluorescence microscopy (Nikon A1R).
Airway hyperresponsiveness
Airway hyperresponsiveness was assessed as previously described [17]. Briefly, mice were intranasally administered 2 µg of α-Galcer in 50 µl of PBS. After 24 h, the mice were anesthetized. A tracheal cannula was inserted and connected to a computer-controlled ventilator. Airway resistance was measured after administration of increasing concentrations of methacholine (10, 25, 50, and 100 mg/ml) in 0.9% NaCl through a nebulizer. Airway resistance was then measured and graphed (Lung resistance cmH2O/mL/s).
ELISA
The levels of serum IL-4 were measured by using an ELISA Kit (eBioscience) according to the manufacturer’s protocols.
Statistical analysis
Statistical significance was assessed by two-tailed unpaired Student’s t tests with Prism 7 software, (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001), unless otherwise stated.
Results
PGRN deficiency impairs the development of iNKT cells
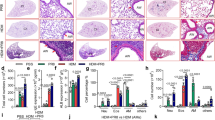
To investigate the role of PGRN in iNKT cell development, we compared the percentage and total number of thymic iNKT cells between PGRN KO and WT mice on the same background. Using CD1d and TCR-β staining, the percentage of iNKT cells was higher but the absolute cell number was lower in PGRN KO mice than in WT controls (Fig. 1A, B). iNKT cells arise from double positive thymocytes and are divided into four populations (stage 0 to stage 3) [18]. CD1d, CD24, CD44, and NK1.1 were used as markers to determine the iNKT cell stage [18]. We found that the percentage and absolute number of stage 0 (CD1d+CD24+) and stage 1 (CD44-NK1.1-) cells were decreased in the PGRN KO cells, but these values for stage 2 (CD44+NK1.1-) and stage 3 (CD44+NK1.1+) cells were not significantly different between WT and KO mice (Fig. 1C–E). To determine whether the development of PGRN KO iNKT cells was cell-intrinsic, we generated bone marrow chimeric mice by transferring CD45.2 WT or PGRN KO:CD45.1 WT bone marrow cells at 1:1 to WT CD45.1 recipient mice. After 8 weeks, the percentage of CD45.2 PGRN KO iNKT cells was found to be higher than that of the CD45.2 WT iNKT cells (Fig. 1F, H). The ratio of CD45.2 PGRN KO iNKT cells in stage 0 and stage 1 was also decreased in the chimeric mice, but there was no difference in the stage 2 and stage 3 iNKT cells (Fig. 1G, I). These data show that the impact of PGRN on iNKT development is cell-autonomous. We also quantified splenic iNKT cells from the WT and PGRN KO mice, and found that the percentage of iNKT cells was significantly increased in the KO mice, but the absolute cell number was not significantly different (Fig. S1A-B). Furthermore, we found that PGRN expression was higher in stages 0 and 1 than in stages 2 and 3 (Fig. 1J). These findings indicate that PGRN is important for stage 0 and stage 1 iNKT cell development and differentiation.
A, B Flow cytometry data of iNKT cells from WT and PGRN KO mice. Unloaded CD1d was used as a negative control (n = 11). C–E Flow cytometry data of stage 0 to stage 3 cells from WT and PGRN KO mice (n = 9). F Flow cytometry data of iNKT cells from WT and PGRN KO chimeric mice. G Flow cytometry data of stage 0 to stage 3 cells from WT and PGRN KO chimeric mice. H Percentage of cell populations gated from (F) (n = 3). I Percentage of cell populations gated from (G) (n = 3). J PGRN expression in stage 0 to stage 3 iNKT cells from WT mice. Flow diagrams are representative of 3 independent experiments. Data are presented as means ± SEMs. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
PGRN deficiency results in impaired expansion of NKT2 cells
iNKT cells can be classified into NKT1, NKT2, and NKT17 cells based on differential expression of transcriptional factors that produce IFN-γ, IL-4, and IL-17, respectively [19]. To investigate the role of PGRN in the differentiation of NKT1, NKT2, and NKT17 cells, we stained thymic iNKT cells for intracellular expression of PLZF, Tbet, GATA3, and RORγt, and found that the percentage and absolute number of NKT2 cells were decreased in the PGRN KO mice, but there was no obvious difference in NKT17 and NKT1 cells between the WTs and KOs (Fig. 2A, C). We also utilized the chimeric mice to investigate whether the change in NKT2 in the PGRN KOs was cell-intrinsic. Consistently, the results showed that the percentage of NKT2 cells in the CD45.2 PGRN KO mice was also decreased, but the NKT1 and NKT17 cell percentages were not different between the WT and PGRN KO mice (Fig. 2B, D). These results indicate that the change in NKT2 cells in the KO mice was cell-intrinsic. Furthermore, we found the splenic NKT2 cells was reduced in the PGRN KO mice, but NKT1 and NKT17 cells have no change between WTs and KOs (Fig. S1C, E). We also detected PGRN expression in different iNKT cell subsets from WT mice and found that PGRN was expressed the highest in NKT2 cells (Fig. 2E). All these results suggest that PGRN is important for NKT2 cell differentiation.
A Flow cytometry data of NKT2, NKT17, and NKT1 cells from WT and PGRN KO mice. B Flow cytometry data of NKT2, NKT17, and NKT1 cells from WT and PGRN KO chimeric mice. C Percentage and absolute number of cell populations gated from (A) (n = 10). D Percentage of cell populations gated from (B) (n = 3). E PGRN expression in NKT1, NKT2, and NKT17 cells from WT mice. Data are presented as means ± SEMs. ∗p < 0.05; ∗∗∗p < 0.001.
PGRN is critical for NKT2 cell effector function
NKT2 cells produce IL-4 and play an important role in certain diseases such as allergic asthma-like lung pathology observed in mice [20]. We stimulated thymic and splenic iNKT cells from WT and PGRN KO mice with PMA and ionomycin in vitro for 5 h and examined cytokine production by flow cytometry. Compared to the corresponding WT cells, IL-4 production was decreased in both PGRN KO thymic and splenic iNKT cells, TNF-α expression was increased in PGRN KO thymic iNKT cells, and IL-17 and IFN-γ were not significantly different (Fig. 3A, C, Fig. S1D, F). By using chimeric mice, we found that the decrease in IL-4 production in the thymus of PGRN KO mice was cell-intrinsic (Fig. 3B, D). To further confirm the effects of PGRN on iNKT cell cytokine production under physiological conditions, thymic iNKT cells were stimulated with α-Galcer in vivo and in vitro. As assessed by flow cytometry, the production of IL-4 was decreased but that of IL-17, TNF-α, and IFN-γ was unchanged in the PGRN KO mice (Fig. 3E, H), indicating that PGRN is critical for α-Galcer-induced IL-4 production. Furthermore, we detected the mRNA expression of Il-4 in thymic iNKT cells and found that it was also decreased in the KO mice (Fig. 3F). Because IL-4 produced by iNKT cells was previously shown to induce allergic asthma-like lung pathology in mice [20], we measured IL-4 serum levels in mice after stimulating with α-Galcer and methacholine, and found the levels to be lower in PGRN KO mice than in WT mice (Fig. 3G). We then tested airway resistance after stimulation with aerosolized methacholine. We found that baseline hyperresponsiveness between WT and PGRN KO mice was comparable and both mouse genotypes had elevated airway resistance after being stimulated with aerosolized methacholine. However, PGRN KO mice have decreased airway resistance compared with WT controls (Fig. 3I). These results indicate that PGRN deficiency results in impaired function of NKT2 cells.
A Enriched thymus iNKT cells were stimulated with PMA and ionomycin in vitro for 5 h and the expression of IL-4, IL-17, TNF-α, and IFN-γ in enriched iNKT cells was analyzed by flow cytometry. B Flow cytometry analysis of IL-4, IL-17, TNF-α, and IFN-γ cells in enriched iNKT cells from WT and PGRN KO chimeric mice after stimulation with PMA and ionomycin. C Percentages of selected cell populations from (A), (n = 8). D Percentages of selected cell populations from (B), (n = 3). E Ratio of IL-4, IL-17, TNF-α, and IFN-γ cells in enriched thymus iNKT cells stimulated with α-Galcer and PMA + ionomycin. F mRNA expression of Il-4 in thymus iNKT cells (n = 5). G Serum titers of IL-4 after treatment with α-Galcer or methacholine in vivo (n = 5). H Percentages of selected cell populations from (E) (n = 3). I Airway resistance data of WT and PGRN KO mice after stimulation with methacholine (n = 4). Data are presented as means ± SEMs. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Loss of PGRN increases apoptosis of stage 1 iNKT cells
We investigated whether the reduction in thymic iNKT and NKT2 cells in PGRN KO mice was caused by changes in proliferation or apoptosis. First, we examined the proliferation of iNKT cells by flow cytometry and found no difference in the percentage of Ki67+ NKT1, NKT2, NKT17, and total iNKT cells between WT and PGRN KO mice (Fig. 4A, B). We also found no change in the proportion of Ki67+ cells for CD45.2 PGRN KO iNKT, NKT1, NKT2, and NKT17 cells (Fig. 4C, D). We then examined the expression of Annexin V in different stages of iNKT cells and found that the proportion of Annexin V+ cells was increased in the stage 1 iNKT cells of PGRN KO mice, the majority of which were NKT2 cells. There was no observed difference in Annexin V+ total iNKT and stage 0, 2, 3 iNKT cells (Fig. 4E, F). Furthermore, we found the percentage of Annexin V+ cells to be higher in CD45.2 PGRN KO mice than in WT controls. This indicates that the elevation of Annexin V in stage 1 iNKT cells was cell-intrinsic (Fig. 4G, H) and probably accounted for the reduction in NKT2 cell numbers. We thus conclude that loss of PGRN increases the apoptosis of stage 1 iNKT cells.
A, B Ratio of Ki67+ cells in iNKT, NKT1, NKT2, and NKT17 cells from WT and PGRN KO mice (n = 6). C, D Ratio of Ki67+ cells in iNKT, NKT1, NKT2, and NKT17 cells from chimeric mice (n = 3). E, F Flow cytometry analysis of Annexin V+ cells in iNKT, stage 0, stage 1, stage 2, and stage 3 cells from WT and PGRN KO mice (n = 6). G, H Flow cytometry analysis of Annexin V+ cells in iNKT, stage 0, stage 1, stage 2, and stage 3 cells from chimeric mice (n = 3). Data are presented as means ± SEMs. ∗p < 0.05; ∗∗p < 0.01.
PGRN regulates differentiation of NKT2 cells by promoting expression of PLZF
Next, we investigated how PGRN deficiency leads to the decrease in NKT2 cells. Several transcription factors have been identified that affect NKT2 cell development. Notably, GATA3 is the main transcription factor of NKT2 cells [21], strong TCR signaling induces high PLZF expression that is required for NKT2 differentiation [22], EGR2 directly binds to the promoter of Zbtb16 and promotes PLZF expression [23], and IRF4 promotes IL-4 production by activating the Il-4 promoter [24]. We first examined the expression of Egr2, Irf4, and Zbtb16 mRNA by RT-PCR and found no differences between WT and PGRN KO mice (Fig. 5A–C). Next, as assessed by flow cytometry, the expression of GATA3, RORγt, and Tbet was comparable between WT and PGRN KO mice, but that of PLZF was obviously decreased in the KOs (Fig. 5D). Then we detected the expression of PLZF in CD4+T cells by Western blot, and found that the levels of PLZF in CD4+ T cell lysates were lower in PGRN KO mice than in WT mice (Fig. 5E) (Uncropped Western blots were showed in Fig. S2A). Furthermore, we found that the PLZF+ NK1.1− cell numbers were significantly lower in the PGRN KO mice than in the WT controls (Fig. 5F, G). These results indicate that the KO mice have reduced PLZF expression, which leads to loss of NKT2 cells. In addition, PLZF nuclear localization is important for the development and function of iNKT cells [25]. Thus, we tested whether PGRN affects PLZF nuclear localization by CFm. The results showed that PLZF was localized to both the nucleus and cytoplasm of iNKT cells and the mean fluorescence intensity(MFI) of nuclear PLZF was significantly lower in the KO mice than in the WT controls. After stimulation with α-Galcer for 3 days, we found that PLZF nuclear localization was elevated in the iNKT cells from both genotypes, but the nuclear PLZF MFI ratio was lower in the KO mice than in the WT controls (Fig. 5H–J). All of these results indicate that PGRN regulates NKT2 differentiation and function by promoting the expression and nuclear entry of PLZF.
A Egr2 mRNA expression in iNKT cells from thymuses of WT and PGRN KO mice (n = 5). B Irf4 mRNA expression in iNKT cells from thymuses of WT and PGRN KO mice (n = 5). C Zbtb16 mRNA expression in iNKT cells from thymuses of WT and PGRN KO mice (n = 5). Data are presented as means ± SEMs for (A–C). D Flow cytometry analysis of PLZF, Tbet, RORγt, and GATA3 expression in iNKT cells from thymuses of WT and PGRN KO mice (n = 5). MFI values were analyzed by FlowJo software. E Lysates of CD4+ cells from the thymus of WT and PGRN KO mice were immunoblotted for PLZF, β-actin was used as a loading control. Images are representative of 3 independent experiments. F Flow cytometry data of NK1.1-PLZF+ iNKT cells from thymuses of WT and PGRN KO mice. G Percentage and cell number of the gated population from (F), (n = 4). H–I Sorted iNKT cells from thymuses of WT and PGRN KO mice were incubated with or without 125 ng/ml α-Galcer for 72 h and stained for PLZF. Cells were analyzed using confocal microscopy and the MFI of PLZF was measured. Data are representative images from 3 independent experiments and the means ± SDs from more than 50 cells are plotted. Scale bars, 2.5 μm. J The ratio of nuclear PLZF MFI in iNKT cells before and after stimulation was plotted (n = 3). *p < 0.05; **p < 0.01. Data were analyzed by two-way ANOVA in (I).
PGRN couples with EZH2 to regulate expression and stability of PLZF
The inconsistencies in mRNA and protein expression levels led us to speculate whether the decreased expression of PLZF was caused by abnormalities in post-transcriptional modification. EZH2 is a histone methyltransferase that methylates both H3K27 and non-histone proteins such as transcription factors that play an important role in immune system development and stability [13]. Previous studies have found that EZH2 deficiency in T cells leads to expansion of PLZFhigh NKT cells, which is attributed to PLZF methylation and degradation [26]. Here, we investigated whether the decrease in PLZF in PGRN KO mice correlates with EZH2 expression. First, we examined the mRNA expression of Ezh2 in iNKT cells and found that Ezh2 mRNA levels were significantly higher in PGRN KO mice than in WT mice (Fig. 6A). Next, we analyzed EZH2 protein expression by immunofluorescence using confocal microscopy and found that it was increased in the KO mice (Fig. 6B, C). Furthermore, it was found that the levels of EZH2 in CD4+ T cell lysates were higher in PGRN KO mice than in WT mice (Fig. 6D) (Uncropped Western blots were showed in Fig. S2B). These results indicate that the reduced NKT2 cell numbers and PLZF levels in the PGRN KO mice may be caused by enhanced EZH2 expression.
A Ezh2 mRNA expression in thymus iNKT cells from WT and PGRN KO mice (n = 5). B, C Sorted iNKT cells from thymus were stained for EZH2 and analyzed using confocal microscopy to measure EZH2 MFI. Scale bars, 2.5 μm. **p < 0.01 by Mann–Whitney U test in (C). D Lysates of CD4+ cells from the thymus of WT and PGRN KO mice were immunoblotted for EZH2, β-actin was used as a loading control. Images are representative of 3 independent experiments. E Luciferase assay for pGL3-mEZH2 promoter-luciferase after pGL3-mEZH2 promoter-luciferase and pAd5-E1-CMV or pAd5-E1-CMV-mPGRN were co-transfected into HEK293T cells. Luciferase assay for pGL3-mPLZF promoter-luciferase after pGL3-mPLZF promoter-luciferase and pAd5-E1-CMV or pAd5-E1-CMV-mEZH2-Flag or pAd5-E1-CMV-mPGRN-Flag were co-transfected with into HEK293T cells. Luciferase activities were measured after 48 h of culture (n = 3 individual experiments). F Thymocytes from WT and PGRN KO mice were labeled with anti-CD4, then stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml) for the indicated times and stained for phospho-IKKα/β and phospho-P65. MFI values were analyzed by FlowJo software. G Phosphorylation of P65 and IKKα/β in iNKT cells from WT and PGRN KO mice. H iNKT cells were isolated from WT mice, after stimulation with or without anti-PGRN antibodies, and then stained for EZH2. The expression levels of EZH2 were analyzed by flow cytometry. MFI values were analyzed by FlowJo software. I–N PGRN KO mice were treated with intraperitoneal GSK343 consecutively for 14 days. Shown are the flow cytometry data of iNKT (I) and NKT2 (K) cells from thymocytes of WT and PGRN KO mice, and the frequency and cell number in (J) (gated from (I)), L(gated from (K)), (n = 4). M, N PGRN KO mice were consecutively treated with GSK343 for 14 days. Flow cytometry data of IL-4, IL-17, TNF-α, and IFN-γ expressing cells in enriched iNKT cells after stimulation with PMA and ionomycin for 5 h (n = 4). (O) PGRN KO mice were consecutively treated with GSK343 for 14 days. PLZF expression in iNKT cells of thymuses was analyzed by flow cytometry (n = 4). P Lung resistance of WT, PGRN KO, WT + GSK343, and KO + GSK343 mice after stimulation with methacholine (n = 3). Data were analyzed by two-tailed unpaired Student’s t tests (A), one-way ANOVA (E, J, L, N). Data are presented as means ± SEMs. *p < 0.05; **p < 0.01.
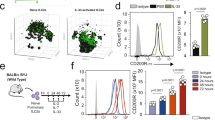
To study how PGRN regulates the expression of Ezh2 or Plzf and EZH2 regulates the expression of Plzf, we carried out a dual luciferase report assays. Firstly, pGL3-mPLZF promoter-luciferase and pAd5-E1-CMV-mEZH2-Flag or pAd5-E1-CMV-mRGRN were co-transfected into HEK293T cells. After 48 h, cells were collected and the luciferase activity of Plzf promoter was tested according to dual-Luciferase ®Report Assay Systems kit. The results showed that PGRN overexpression increased the luciferase activity of Plzf promoter, in contrast, EZH2 overexpression decreased the luciferase activity of Plzf promoter (Fig. 6E). Meanwhile, pGL3-Ezh2 promoter-luciferase and pAd5-E1-CMV-PGRN were co-transfected into HEK293T cells according to the above method. The results were shown that PGRN overexpression decreased the luciferase activity of Ezh2 promoter (Fig. 6E). These results indicated that PGRN promotes the transcription of Plzf and inhibits the transcription of Ezh2, while EZH2 inhibits the transcription of Plzf. Previous studies have shown that depletion of TNF-α inhibits EZH2 expression, which is related to the NF-κB pathway [27]. We asked whether PGRN binds to TNFR to promote EZH2 expression. Thymocytes were labeled with anti-CD4 antibody and stimulated with PMA and ionomycin in vitro, we found that the phosphorylation of IKKα/β and P65, which occurs downstream of TNF-α-TNFR in the NF-κB signaling pathway, was significantly increased in PGRN KO mice compared to that in WT mice (Fig. 6F). We also detected the phosphorylation of P65 and IKKα/β in thymus iNKT cells from WT and PGRN KO mice, and found both to be increased in the latter mice (Fig. 6G). This result suggests that PGRN regulates EZH2 expression through the NF-κB pathway. Furthermore, using anti-PGRN immunofluorescence, we found PGRN localized to both the cell membrane and cytoplasm of iNKT cells (Fig. S1G). We then activated WT iNKT cells with anti-PGRN antibodies and found that EZH2 expression was downregulated (Fig. 6H), indicating that activation of PGRN decreases EZH2 expression. To further confirm that PGRN couples with EZH2 to regulate PLZF expression and stability, PGRN KO mice were treated with the EZH2 inhibitor GSK343 in vivo for 14 consecutive days. We found that the iNKT, NKT2, and IL-4+ cells were rescued by this treatment to the same level as that of WT mice (Fig. 6I–N). In addition, we found that PLZF expression and hyperresponsiveness were ameliorated to WT levels after treatment with GSK343 (Fig. 6O–P). All these results indicate that PGRN couples with EZH2 to regulate the expression and stability of PLZF, which further regulates the differentiation of NKT2 cells.
Discussion
PGRN induces iNKT cells to produce Th2 cytokines, but the underlying molecular mechanism is not clear. In our study, we investigated the role of PGRN in iNKT cells using PGRN KO mice. Compared with WT controls, PGRN KO mice had increased iNKT cell frequency but decreased cell numbers. Interestingly, we found that stage 1 iNKT and NKT2 cells were significantly decreased in the KO mice, while NKT1 and NKT17 cell numbers were unchanged. Using chimeric mice, we demonstrated that the change in iNKT and NKT2 cells was cell-intrinsic. Furthermore, we found that NKT2 cell function was impaired in the KO mice. Mechanistically, the decrease in NKT2 cell numbers correlated with increased expression of EZH2, which in turn caused increased degradation and altered nuclear localization of PLZF, and this was modulated by hyperactivation of TNF-α-TNFR2-mediated pathways. By using luciferase reporter assays, we have proved that PGRN promotes the transcription of Plzf and inhibits the transcription of Ezh2, while EZH2 inhibits the transcription of Plzf. To the best of our knowledge, we have identified a new pathway for PGRN to regulate iNKT cell differentiation and function.
Although PGRN has been widely studied in autoimmune disorders, cancer, and neurodegenerative diseases [28,29,30,31], few studies have focused on immune cells. PGRN has been found to bind TLR9 and it plays an important role in innate immunity [32]. One study indicated that PGRN promotes CD4+ T cells to differentiate into Treg cells, enhancing the latter’s function [6]. PGRN is also highly expressed in a neutrophil subpopulation that promotes antibody diversity in B cells [33]. In addition, PGRN has been shown to induce expression of the Th2-like cytokines, IL-4 and IL-5 [34]. As IL-4 is produced in both iNKT cells and activated CD4+ T cells, we first wanted to uncover the role of PGRN in iNKT cells and the mechanism of PGRN-dependent regulation of iNKT cell development and function. IL-4 is regulated upstream by several transcription factors, including GATA3, IRF4, and c-MAF [35,36,37]. Our study has established another upstream regulation axis (PGRN-EZH2-PLZF) to epigenetically control IL-4 expression, which may be utilized by other cell types in addition to iNKT cells. Thus, we first established the link that PGRN signaling is negatively associated with EZH2 expression.
Previously, NKT0 cells from stage 0 were shown to develop into NKT2 cells, and then NKT1 and NKT17 cells [38]. In our study, we have proved the cell number of stage 0 iNKT cells was reduced in the PGRN KO mice, which may have attenuated the differentiation of NKT2 cells. However, how PGRN influences the generation of stage 0 iNKT cells is still unclear. We found increased NF-κB signaling in the KO mice, which indicates that NF-κB signaling may influence early iNKT cell differentiation. PLZF is a key factor for regulation of iNKT cell development and function, and deletion of PLZF in mice prevents the transition of iNKT cells from stage 1 to stage 2. Moreover, PLZF deficiency in iNKT cells prevents the secretion of high levels of IL-4 and IFN-γ [39, 40]. In our study, we found that stage 0 and stage 1 iNKT cells were significantly decreased with PGRN KO, but there was no change in stage 2 and stage 3 iNKT cell numbers. Furthermore, we found increased levels of Annexin V in stage 1 cells, indicating that the change in NKT2 cells resulted from increased apoptosis and decreased PLZF expression. A previous study revealed that PLZF had an anti-apoptotic effect in many cell types (e.g. Jurkat cells) [41]. In our study, we found that downregulation of PLZF leads to increased NKT2 cell apoptosis, indicating that PLZF has similar anti-apoptotic activity in iNKT cells. Moreover, we found no obvious difference in the IFN-γ produced by activated iNKT cells between WT and PGRN KO mice, which could be caused by residual expression of PLZF in PGRN KO iNKT cells. However, changes in the expression of other proteins because of the absence of PGRN may also be responsible, and other cells that produce IFN-γ may be able to compensate.
High EZH2 expression is observed in a wide range of cancers, including B cell and T cell lymphoid malignancies [42]. Many dysregulated signaling pathways can also lead to abnormal EZH2 expression. MYC upregulates EZH2 through repression of miRNA-26a and miRNA-26b [43]. Downregulation of miRNA-138 leads to increased Ezh2 gene targeting [44]. The NF-κB transcription factor Rel and IL-6 upregulate the expression of EZH2 in T-ALL and multiple myeloma cells [45], and depletion of PI3K/AKT decreases EZH2 expression. In our study, we found that the phosphorylation of S6 ribosomal protein and AKT in iNKT cells upon stimulation was not different in PGRN KO and WT mice (data not shown). However, NF-κB signaling was upregulated in PGRN KO iNKT cells upon stimulation. Therefore, it is highly possible that PGRN signaling inhibits the activation of NF-κB, which in turn leads to EZH2 downregulation. The decreased EZH2 expression upregulates PLZF, and altogether these increase NKT2 and Th2 cell responses. Moreover, luciferase reporter assays were conducted to demonstrate the interactions among Pgrn, Plzf and Ezh2. As a result, PGRN positively regulates the Plzf promoter activity, but negatively regulates the Ezh2 promoter activity. EZH2 negatively regulates the Plzf promoter activity. Thus, we have established a regulatory network centralized on EZH2 regulating NKT2 cell differentiation as well as Th2 responses. The high expression of EZH2 in PGRN KO mice may be the result of increased TNF-α-TNFR signaling.
In conclusion, PGRN regulates the development and function of NKT2 cells via downregulating EZH2 expression and upregulating PLZF expression. This is a novel mechanism of how PGRN links to airway hyperactivity in mice and could be a new target for the treatment of asthma.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Pogonowska M, Poniatowski LA, Wawrzyniak A, Krolikowska K, Kalicki B. The role of progranulin (PGRN) in the modulation of anti-inflammatory response in asthma. Cent Eur J Immunol. 2019;44:97–101.
Finkelman FD, Hogan SP, Hershey GK, Rothenberg ME, Wills-Karp M. Importance of cytokines in murine allergic airway disease and human asthma. J Immunol (Baltim, Md: 1950). 2010;184:1663–74.
Lisbonne M, Diem S, de Castro Keller A, Lefort J, Araujo LM, Hachem P, et al. Cutting edge: invariant V alpha 14 NKT cells are required for allergen-induced airway inflammation and hyperreactivity in an experimental asthma model. J Immunol (Baltim, Md: 1950). 2003;171:1637–41.
Reynolds C, Barkans J, Clark P, Kariyawasam H, Altmann D, Kay B, et al. Natural killer T cells in bronchial biopsies from human allergen challenge model of allergic asthma. J Allergy Clin Immunol. 2009;124:860–2.
Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48:999–1009.
Tang W, Lu Y, Tian QY, Zhang Y, Guo FJ, Liu GY, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–84.
Wei F, Zhang Y, Jian J, Mundra JJ, Tian Q, Lin J, et al. PGRN protects against colitis progression in mice in an IL-10 and TNFR2 dependent manner. Sci Rep. 2014;4:7023.
Park SY, Hong GH, Park S, Shin B, Yoon SY, Kwon HS, et al. Serum progranulin as an indicator of neutrophilic airway inflammation and asthma severity. Ann Allergy Asthma Immunol. 2016;117:646–50.
Choi JP, Park SY, Moon KA, Ha EH, Woo YD, Chung DH, et al. Macrophage-derived progranulin promotes allergen-induced airway inflammation. Allergy. 2020;75:1133–45.
Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96.
Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–36.
Gunawan M, Venkatesan N, Loh JT, Wong JF, Berger H, Neo WH, et al. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol. 2015;16:505–16.
Tumes D, Hirahara K, Papadopoulos M, Shinoda K, Onodera A, Kumagai J, et al. Ezh2 controls development of natural killer T cells, which cause spontaneous asthma-like pathology. J Allergy Clin Immunol. 2019;144:549–560. e510.
Yang K, Blanco DB, Neale G, Vogel P, Avila J, Clish CB, et al. Homeostatic control of metabolic and functional fitness of T reg cells by LKB1 signalling. Nature. 2017;548:602–6. %@ 1476-4687
Zhu K, Wang Y, Liu L, Li S, Yu W. Long non-coding RNA MBNL1-AS1 regulates proliferation, migration, and invasion of cancer stem cells in colon cancer by interacting with MYL9 via sponging microRNA-412-3p. Clin Res Hepatol Gastroenterol. 2020;44:101–14.
Du Z, Yang D, Zhang Y, Xuan X, Liu C. AKT2 deficiency impairs formation of the BCR signalosome. Cell Commun Signal. 2020;18:56.
Wu J, Yang J, Yang K, Wang H, Gorentla B, Shin J, et al. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J Clin Investig. 2014;124:1685.
Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336.
Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–54.
Akbari, O, Stock, P, Meyer, E, Kronenberg M, Sidobre S, Nakayama T, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003;9:582–8.
Zhang X, Wang K, Zhao W, Cao L, Zhang S, Jin R, et al. TRAF3IP3 at the trans-Golgi network regulates NKT2 maturation via the MEK/ERK signaling pathway. Cell Mol Immunol. 2020;17:395–406. %@ 2042-0226
Tuttle KD, Krovi SH, Zhang J, Bedel R, Harmacek L, Peterson LK, et al. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat Commun. 2018;9:2650.
Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13:264–71.
Tominaga N, Ohkusu-Tsukada K, Udono H, Abe R, Matsuyama T, Yui K. Development of Th1 and not Th2 immune responses in mice lacking IFN-regulatory factor-4. Int Immunol. 2003;15:1–10.
Zhang L, Tschumi BO, Corgnac S, Ruegg MA, Hall MN, Mach JP, et al. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. J Immunol (Baltim, Md: 1950). 2014;193:1759–65.
Vasanthakumar A, Xu D, Lun AT, Kueh AJ, van Gisbergen KP, Iannarella N, et al. A non-canonical function of Ezh2 preserves immune homeostasis. EMBO Rep. 2017;18:619–31.
Urbano PCM, Koenen H, Joosten I, He X. An autocrine TNFalpha-tumor necrosis factor receptor 2 loop promotes epigenetic effects inducing human treg stability in vitro. Front Immunol. 2018;9:573.
Wei J, Hettinghouse A, Liu C. The role of progranulin in arthritis. Ann NY Acad Sci. 2016;1383:5–20.
Tian G, Jin X, Wang Q, Ye T, Li G, Liu J. Recent advances in the study of progranulin and its role in sepsis. Int Immunopharmacol. 2019;79:106090.
Cui Y, Hettinghouse A, Liu CJ. Progranulin: a conductor of receptors orchestra, a chaperone of lysosomal enzymes and a therapeutic target for multiple diseases. Cytokine Growth Factor Rev. 2019;45:53–64.
Mendsaikhan A, Tooyama I, Walker DG. Microglial progranulin: involvement in Alzheimer’s Disease and neurodegenerative diseases. Cells. 2019;8:230.
Park B, Buti L, Lee S, Matsuwaki T, Spooner E, Brinkmann MM, et al. Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity. 2011;34:505–13.
Puga I, Cols M, Barra CM, He B, Cassis L, Gentile M, et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat Immunol. 2011;13:170–80.
Pickford F, Marcus J, Camargo LM, Xiao Q, Graham D, Mo JR, et al. Progranulin is a chemoattractant for microglia and stimulates their endocytic activity. Am J Pathol. 2011;178:284–95.
Hu CM, Jang SY, Fanzo JC, Pernis AB. Modulation of T cell cytokine production by interferon regulatory factor-4. J Biol Chem. 2002;277:49238–46.
Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96.
I C H. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–83.
Baranek T, Lebrigand K, de Amat Herbozo C, Gonzalez L, Bogard G, Dietrich C, et al. High dimensional single-cell analysis reveals iNKT cell developmental trajectories and effector fate decision. Cell Rep. 2020;32:108116.
Mao AP, Constantinides MG, Mathew R, Zuo Z, Chen X, Weirauch MT, et al. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci USA. 2016;113:7602–7.
Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403.
Parrado A, Robledo M, Moya-Quiles MR, Marin LA, Chomienne C, Padua RA, et al. The promyelocytic leukemia zinc finger protein down-regulates apoptosis and expression of the proapoptotic BID protein in lymphocytes. Proc Natl Acad Sci USA. 2004;101:1898–903.
Li B, Chng W-J. EZH2 abnormalities in lymphoid malignancies: underlying mechanisms and therapeutic implications. J Hematol Oncol. 2019;12:1–13. 1756-8722
Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–12.
Rastgoo N, Pourabdollah M, Abdi J, Reece D, Chang H. Dysregulation of EZH2/miR-138 axis contributes to drug resistance in multiple myeloma by downregulating RBPMS. Leukemia. 2018;32:2471–82.
Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–80.
Funding
This study was supported by the National Natural Science Foundation of China (32100710).
Author information
Authors and Affiliations
Contributions
ZCD carried out the initial analyses and drafted the initial manuscript. ZCD, DY, LH, LLN, CSR, HL, LLH and LJZ performed the flow cytometry assay. ZCD, JS, XQS performed the immunoblotting, confocal microscopy, RT-PCR, and ELISA. ZCD and DJ performed the airway hyperresponsiveness. XD performed the plasmid construction and luciferase reporter assay. HM, PH and YC reviewed and revised the manuscript. XDZ and CHL conceptualized and designed the study. Thanks for the discussion with Professor You Sook Cho about our work. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal work was reviewed and approved by the Institutional Animal Care and Use Committee of the Children’s Hospital of Chongqing Medical University and all experiments were performed according to the principles of medical ethics and the requirements of the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Y Shi
Rights and permissions
About this article
Cite this article
Du, Z., Huang, L., Dai, X. et al. Progranulin regulates the development and function of NKT2 cells through EZH2 and PLZF. Cell Death Differ 29, 1901–1912 (2022). https://doi.org/10.1038/s41418-022-00973-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-022-00973-6