Abstract
The Atlantic Ocean-Mediterranean Sea junction has been proposed as an important phylogeographical area on the basis of concordance in genetic patterns observed at allozyme, mtDNA and microsatellite DNA markers in several marine species. This study presents microsatellite DNA data for a mobile invertebrate species in this area, the cuttlefish Sepia officinalis, allowing comparison of this relatively new class of DNA marker with previous allozyme results, and examination of the relative effects on gene flow of the Strait of Gibraltar and the Almería-Oran oceanographic front. Genetic variation at seven microsatellite loci screened in six samples from NE Atlantic and Mediterranean coasts of the Iberian Peninsula was high (mean Na = 9.6, mean He = 0.725). Microsatellites detected highly significant subpopulation structuring (FST= 0.061; RST = 0.104), consistent with an isolation-by-distance model of low levels of gene flow. Distinct and significant clinal changes in allele frequencies between Atlantic and Mediterranean samples found at five out of seven loci, however indicate these results might be also consistent with an alternative model of secondary contact and introgression between previously isolated and divergent populations, as previously proposed for other marine species from the Atlantic-Mediterranean area. A pronounced ‘step’ change between SW Mediterranean samples associated with the Almería-Oran front suggests this oceanographic feature may represent a contemporary barrier to gene flow.
Similar content being viewed by others
Introduction
Studies on several fishes, molluscs and crustaceans (see Sanjuan et al, 1996, 1997 and references therein; Roldán et al, 1998; Naciri et al, 1999; Zane et al, 2000) have suggested the importance of the junction of the Atlantic Ocean and Mediterranean Sea in phylogeographical structuring of marine species. To explain the concordance of patterns of genetic differentiation observed in these marine species, a hypothesis has been proposed wherein Mediterranean and Atlantic populations have been isolated due to sea level changes during glacial maxima and subsequently reconnected, possibly several times, in the past. Genetic divergence of populations during isolation followed by secondary contact after the re-opening of the Strait of Gibraltar, and with currently restricted gene flow between the geographically divergent populations (ie, secondary intergradation), are predicted to have produced present patterns in genetic structuring. Two factors suggested to account for the contemporary position and maintenance of discontinuities in frequencies of genetic markers in populations in this area are one-way surface current flows through the Strait of Gibraltar (eg, allozymes in hake – Roldán et al, 1998) and the presence of a well defined hydrographic boundary of surface waters between Almería (SE Spain) and Oran (Algeria) (Tintore et al, 1988), the Almería-Oran oceanographic front (eg, allozymes and mtDNA in mussels – Quesada et al, 1995; Sanjuan et al, 1996; microsatellites in fish – Naciri et al, 1999). Nevertheless, the possibility of such geographically based genetic patterns resulting from adaptive responses to different environmental pressures (natural selection) or due to isolation by distance among populations, cannot presently be excluded.
Recently Pérez-Losada et al (1999) described highly significant subpopulation structuring and clinal allozyme frequency changes between Atlantic and Mediterranean samples of the cuttlefish Sepia officinalis around the Iberian peninsula, which they explained on the basis of the secondary intergradation model described above. Their results did not however give any clear indication of a distinct genetic change between samples from either side of the Strait of Gibraltar or the Almería-Oran front. Dispersal ability is presumed to be restricted in S. officinalis: females fix their eggs to the sea floor, a pelagic larval phase is lacking (benthic juveniles hatch direct from eggs), and the adults have limited migratory capacities (Guerra, 1992). The Almería-Oran oceanographic front is associated with steep temperature (1.4°C) and salinity (2 psu) gradients over a distance of 2 km, and with strong water currents (40 cm/s average speed) that flow in a south-easterly direction away from the Spanish coast toward North Africa (Tintore et al, 1988), whilst the Strait of Gibraltar experiences one-way surface current flows (Parilla and Kinder, 1992). Consequently, distinct genetic differentiation between S. officinalis samples to the west and east of these oceanographic features might be expected.
The aim of the present study was to use microsatellite DNA markers to: (i) check alternative hypotheses (ie, secondary intergradation, isolation by distance, and natural selection) concerning the genetic structuring of S.officinalis within the Atlantic-Mediterranean area; and (ii) test whether the Strait of Gibraltar or the Almería-Oran front represent a contemporary barrier to extensive gene flow in this species. Microsatellite markers have shown considerable utility for genetic studies of population structure, particularly in organisms exhibiting low variability detectable by other methods (Hughes and Queller, 1993; Estoup et al, 1998). Several recent studies of widespread and abundant marine species (eg, squid, Shaw et al, 1999a; and herring, Shaw et al, 1999b) have indicated the greater capacity of microsatellite vs allozyme markers to detect fine population structure in such species. Microsatellites are also presumed to be neutral markers whereas allozymes have been suggested to be subject to selective forces, a potentially complicating factor when clinal frequency changes are found. The current study reports an application of highly polymorphic microsatellite loci to the question of genetic structuring between Atlantic and Mediterranean populations, and allows comparison of these relatively new molecular markers with data from allozymes.
Methods
Sampling
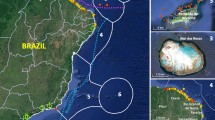
Six samples of S. officinalis were collected from commercial catches landed in Iberian Peninsula fishing ports between July and December 1998 (Figure 1). All catches in this area are taken within short distances (<25 km) of the port of landing, so they can be accepted as reliably representing local S. officinalis populations. Three samples were from the NE Atlantic: Riveira in west Galicia (GalW, n = 66), Lisbon (Lisb, n = 38) and Faro (Faro, n = 70). Three were from the Mediterranean: Roquetas del Mar (RMar, n = 65), south-west of the Almería-Oran oceanographic front; Alicante (Alic, n = 65) and Vilanova i la Geltrú (VGel, n = 63), both north-east of the Almería-Oran front. The specimens were obtained on the date of capture and were immediately frozen in dry ice and transported to the laboratory, where arm tips or mantle tissue were dissected and preserved in absolute ethanol.
Samples of Sepia officinalis collected in fishing ports along the Atlantic and Mediterranean coasts. Sample codes are indicated in Table 1. Arrows indicate the marine surface currents in the SW Mediterranean Sea (from Tintore et al, 1988).
DNA extraction and microsatellite analysis
Total DNA was obtained using a standard proteinase K extraction procedure followed by addition of 5M NaCl and precipitation with isopropanol. All six samples were screened for variation at each of seven polymorphic microsatellite loci (Sof1 to Sof7) previously isolated and characterised for S. officinalis by Shaw and Pérez-Losada (1999). PCR reactions were performed under conditions set out in Shaw and Pérez-Losada (1999). Reaction mixes contained 20 ng template DNA, 1.5–2.5 mM MgCl, 0.2 mM each nucleotide, 0.2 μM of each primer (forward primer 5’ end-labelled with a Cy5 fluorescent dye group), 0.2 U Taq polymerase (Bioline UK) with the manufacturer’s supplied 1x NH4 buffer, in a final reaction volume of 10 μl. Amplified products were resolved on 6% denaturing polyacrylamide gels run on an ALFexpressTM (Pharmacia Biotech) automated sequencer, product sizes being determined by comparison to an internal series of standard size markers using FRAGMENT MANAGER 1.2 software (Pharmacia Biotech).
Differences in product sizes were consistent with all alleles at all loci representing simple three bp repeat motif variation, except for locus Sof3. As described by Shaw and Pérez-Losada (1999) Sof3 displays two parallel sets of allelic arrays differing by one bp: a ‘long’ series of 16 alleles (172–220 bp), and a ‘short’ series of seven alleles (185–203 bp). To simplify the calculation of the various statistics described below at locus Sof3, alleles differing by 1 bp were binned to produce a synthetic array of 16 alleles equivalent to the ‘long’ series (eg, 184 and 185 = 184; 187 and 188 = 187; etc). Comparison of binned, unbinned and adjusted (’short’ and ‘long’ series allele sizes adjusted to create between-series distances much greater than within-series for stepwise mutation model (SMM) statistics) datasets for this locus indicated no substantial differences in any values, and no change in significance, of multi-locus population measures, so binned data are used from here on unless otherwise stated.
Statistical analysis
Genetic polymorphism within samples was measured as the mean number of alleles per locus, and observed and unbiased expected heterozygosity (Nei, 1987). Genotypes at all pairs of loci were tested for genotypic disequilibrium, and deviations from Hardy-Weinberg expectations within loci and over all loci within populations using exact tests with significance determined by a Markov chain method (GENEPOP 3.1b; Raymond and Rousset, 1995).
Differentiation between and genetic relationships among samples can be estimated from microsatellite data using statistics that assume either an infinite alleles model (IAM, Kimura and Crow, 1964) or a stepwise mutation model (SMM, Kimura and Ohta, 1978). As loci Sof3 and Sof5 both display allelic arrays that suggest a non-SMM (Shaw and Pérez-Losada, 1999), non-SMM-based methods (FST, Dchord and Nei’s D) were used preferentially in the following analyses. Nevertheless, because the performance of statistics based on one or another mutation model is still a matter of argument (Estoup et al, 1995; Takezaki and Nei, 1996; Goodman, 1997; Estoup et al, 1998) and for comparative purposes, SMM statistics were also included [RST and (δμ)2]. FST was estimated by θ (Weir and Cockerham, 1984) using F-STAT, and unbiased RST using the RST-CALC programme (Goodman, 1997), which adjusts for differences in sample size and allele size variances among loci. Values for both statistics were tested for significant departure from zero using permutation tests contained within the respective software packages. We also tested for simple frequency differentiation between pairs of samples with Fisher’s exact test, using GENEPOP: multi-locus probabilities were produced using Fisher’s (1954) method for combining probabilities. To test for isolation by distance as an explanation for genetic differences, Mantel tests were performed in GENEPOP (using 1000 permutations to test significance of correlations) on matrices of pairwise geographical distances and θ between samples. The significance of microsatellite variation patterns was also tested by correlation coefficients between the arcsine-squared-root-transformed allele frequencies and geographical distances.
Genetic relationships between Sepia samples were investigated using IAM-based distances: Cavalli-Sforza and Edwards’s (1967) chord distance (Dchord) and Nei’s (1972) distance (D); and the SMM-based distance of Goldstein et al (1995) (δμ)2 which considers allele size differences. These values were then used to construct neighbour-joining (NJ) trees (Saitou and Nei, 1987) using the NJBP programme (A Estoup, J-M Cornuet and S Piry, INRA, Montpelier). Bootstrap support on branches is computed by resampling loci and is given as percentages of 5000 replications.
Results
Variability within samples
Allele sizes and estimates of variability at the seven microsatellite loci screened within all samples are displayed in Table 1. Within-sample variability was uniformly high across all samples: mean number of alleles (Na) ranged between 9.0 and 10.7, and mean observed (Ho) and unbiased expected heterozygosity (He) ranged between 0.567 and 0.700, and 0.614 and 0.784, respectively. Lisbon exhibited a lower number of alleles at loci Sof3, Sof4 and Sof6, although this is likely due to sampling effects (n = 38, compared to mean n = 65 in the other samples). The two samples collected north-east of the Almería-Oran oceanographic front (Alic and VGel) exhibited the lowest levels of heterozygosity, due to substantially lower variability at three loci (Sof1, Sof5 and Sof7) than was present in the samples from west of the front.
Eleven significant departures from outcrossing expectations for genotype frequencies, all due to deficits of heterozygotes, were found in 42 single locus tests (Table 1). Ten tests, associated exclusively with loci Sof4 and Sof7, remained significant when adjusted for table-wide significance by sequential Bonferroni procedure (Rice, 1989). Although five loci therefore suggest close conformity to Hardy-Weinberg proportions in all samples, pooling across all seven loci resulted in highly significant indications of heterozygote deficiencies in each of the six samples.
Exact tests for genotypic disequilibrium between pairs of loci within samples resulted in 11 significant values (P < 0.05) out of 126 comparisons (six significant values are expected at the 5% level). Four significant values out of 21 were observed in the Galician sample, two in Faro, Roquetas and Vilanova, and one in Alicante. All loci, except Sof2 and Sof3, appeared in one or more significant pairwise comparison, although after application of Bonferroni correction only the tests involving Sof1, Sof5 and Sof7 remained significant: Sof1-Sof5 at GalW, RMar, Alic and VGel; Sof1-Sof7 at GalW, Faro and VGel; and Sof5-Sof7 at GalW. It therefore appears that there is no evidence for significant linkage disequilibrium between most of the loci screened here, although the disproportionate number of highly significant tests occurring among the Sof1-Sof5-Sof7 group does indicate that there may be some degree of statistical linkage among these three loci in the samples screened, and especially so in the GalW sample. Genotypic disequilibrium among loci may have significant effects on patterns of variation observed, and in particular it may distort patterns of genetic differences indicated between samples. Therefore, in the following analyses the possible effects of statistical linkage among loci Sof1, Sof5 and Sof7 are evaluated by two means: (i) removing all three loci from the analysis to determine if the four unlinked loci display the same patterns as the total seven-locus dataset; (ii) removing only locus Sof1, as this appears to be the locus generating most of the apparent linkage (over all samples, the Sof5-Sof7 comparison is non-significant if GalW is removed).
Population subdivision
Exact tests for homogeneity of allele frequencies among all samples indicated highly significant differences, both at individual loci and over all loci combined. Similarly the among-sample component of genetic variation, estimated by both FST (θ) and RST, indicated moderate and significant genetic differentiation: θ = 0.061 (P < 0.001); RST = 0.104 (P < 0.001). Pairwise exact tests for multi-locus allele frequency homogeneity indicated highly significant differences between all samples. Recalculation of multi-locus probabilities after removal of the three loci (Sof1, Sof5 and Sof7) showing some sign of linkage disequilibrium, indicated most comparisons were still highly significant (P < 0.001), although two pairwise comparisons were non-significant (GalW-Faro and Alic-VGel) and a further two showed substantially less significant values (GalW-Lisb P = 0.008 and Faro-RMar P = 0.002). Interestingly, of the ‘nearest geographical neighbour’ comparisons only the samples to either side of the Almería-Oran front (RMar-Alic) remain significantly different at the P < 0.001 level. Removal of locus Sof1 only did not change results from those seen for seven-locus comparisons. Similarly, θ and RST showed significant levels of inter-sample genetic variation for the majority of pairwise comparisons (Table 2). Estimates of FST (θ = 0.000–0.179) are consistently lower than equivalent RST values (RST = −0.001–0.292), although oddly RST shows fewer significant comparisons after Bonferroni correction, a result of the greater variance in single locus RST values (despite the RST-CALC correction). Both statistics show lowest values (Table 2) between the geographically closest pairs of samples (ranging between 0.000 and 0.032), except for RMar-Alic where values are clearly higher (0.069 for θ and 0.083 for RST). A pattern of increasing genetic differences with increasing geographic distance between samples is clearly evident, and Mantel tests indicated this to be a highly significant correlation for both θ (P = 0.006) and RST (P = 0.003). Removal of Sof1 did not significantly change pairwise θ and RST values, or significance levels of differences between samples. Increasing genetic differences between samples with increasing geographic distance is not simply due to random changes in allele frequencies accumulating over loci – correlation of frequencies of common alleles (≥10%) showed significant clinal changes from one end of the geographical range to the other for loci Sof1, Sof2, Sof3, Sof5 and Sof7 (after Bonferroni correction). Plots of the most clear examples of clinal frequency changes in common alleles are shown in Figure 2. Interestingly, alleles comprising the two parallel size arrays identified at locus Sof3 also show an array-related clinal distribution (Figure 2) – if all alleles belonging to the ‘long’ series are pooled, the pooled frequency increases from 0.357 off NW Spain (GalW) to 0.817 in the Mediterranean (VGel). Loci Sof4 and Sof6 also exhibited substantial allele frequency changes in this area (see Figure 2), although overall frequency changes did not display significant clinal patterns at these loci. For all five loci showing clinal allele frequency changes, the largest changes occur between the two samples to either side of the Almería-Oran front (RMar and Alic). Alic seems to have diverged more from all the other Atlantic samples and RMar than the more distant sample VGel. This could be explained because of the greater isolation of this sample relative to the others due to its proximity to the front and the oceanographic conditions of the area (ie, Alic can only interchange genes with the samples occurring north of it. See also Sanjuan et al (1996)). RMar, although also close to the front, is favoured by the circulation of the surface currents between SW Mediterranean and north of Africa, which allows an interchange of individuals between both areas (see Figure 1). Nevertheless, other factors such as a reduced historic effective population size and the consequent genetic drift of allele frequencies could also contribute to explain the differentiation of the Alic sample. The low mean heterozygosity values showed by Alic may lend support to this hypothesis.
Geographical variation in frequencies of microsatellite alleles Sof1*220, Sof4*147 (1/n), Sof5*126 and Sof7*171, and in the ‘long’ allele series (l.s.) of locus Sof3 (see text), in Sepia officinalis samples around the Iberian Peninsula. GalW is at 0 km and VGel is at ∼2000 km. A-O front: Almería-Oran oceanographic front. Strait: Strait of Gibraltar.
Hierarchical testing, using WINAMOVA (Excoffier et al, 1992), of samples from different geographical groupings indicated greater genetic heterogeneity among Mediterranean than among NE Atlantic samples, with approximately 6.3% of total variation (θ = 0.067) among samples distributed between the two areas. When samples were grouped into those west and east of the Almería-Oran front, differences within the Mediterranean (east) substantially decreased, but differences between the two areas (east vs west) increase to approximately 7.9% of the total variation (θ = 0.082).
Genetic relationships among samples
Unrooted neighbour-joining trees based on the standard genetic distance of Nei (1972) (D), the chord distance of Cavalli-Sforza and Edwards (1967) (Dchord) and the SMM genetic distance of Goldstein et al (1995) (δμ)2 all exhibited the same topology, as shown for Dchord in Figure 3. The two samples from NE of the Almería-Oran oceanographic front (Alic and VGel) consistently cluster together with 100% bootstrap support, as do the three Atlantic samples (GalW, Lisb and Faro), with the Mediterranean sample to the SW of the front (RMar) consistently falling between these two groupings. The only obvious differences between the tree generated using (δμ)2 and those using D or Dchord were slightly shorter branch lengths and lower bootstrap support on all nodes. Trees generated using distances calculated without Sof1 gave the same topology and similar bootstrap values.
Neighbour-joining tree showing relationships among six samples of Sepia officinalis based on Dchord distances calculated from microsatellite data. Bootstrap support, from 5000 replications, is indicated on branches. Sample codes are indicated in Table 1.
Discussion
Genetic variability within samples
Higher levels of polymorphism were displayed at all seven microsatellite loci, and in all six samples of S. officinalis (Table 1) than those estimated for the six most polymorphic allozyme loci used by Pérez-Losada et al (1999; Na = 2.0 ± 0.3 SD and He = 0.256 ± 0.044 SD). This is not an unexpected result given the proposed higher rates of mutation and improved resolution of microsatellite loci relative to allozyme loci, and agrees well with levels of microsatellite variability reported for squid (Adcock et al, 1999; Shaw et al, 1999a). Levels of allozyme variability reported for cephalopods in general have been very low (mean Na = 1.24, Ho =0.035, He = 0.037 – see Pérez-Losada, 1998), and thus it would appear that the high levels of genetic variability being uncovered at microsatellite loci will now allow more powerful and accurate tests of population genetic parameters in cephalopods.
Significant departures from random out-crossing expectations (Table 1) suggests the presence of non-amplifying (’null’) alleles resulting from mutations in primer sites at loci Sof4 and Sof7. Initial tests on the offspring of several female S. officinalis confirmed this hypothesis (PW Shaw, unpublished data). Redesign of primers and re-screening of individuals will be required in future studies to fully confirm the presence of null alleles.
Analyses of genetic differences among samples including and excluding loci Sof1, Sof5 and Sof7 did not significantly bias patterns in, or estimates of, between- sample differentiation, although inclusion of all three loci does slightly inflate these values. The three loci do not appear to be closely linked physically: cross-combination of primers did not result in amplification between loci, up to the several Kb routinely amplified in standard PCR protocols (PW Shaw, unpublished data); and less than half of tests are significant. One possible cause of genotypic disequilibrium in the current study is that the populations sampled represent a zone of secondary contact and introgression/hybridisation between previously isolated and divergent Atlantic and Mediterranean populations (see below). With limited gene flow and some degree of inbreeding within a substructured population (high FSTvalues among samples), it is possible that association of particular alleles at loci on the same chromosome could be maintained for a number of generations. Resolution of the relationship among the loci described here requires further sampling outside this possible range of secondary contact and controlled breeding studies, both of which are being pursued presently. For the above reasons we have included all data from all loci in the analysis, in the interests of presenting a full dataset and not excluding data until a definitive conclusion can be made on linkage relationships among loci.
Population subdivision and gene flow
Moderate, but highly significant levels of inter-sample differentiation (θ= 0.061 and RST = 0.104) were found for S. officinalis around the Iberian Peninsula. All pairs of samples, even those separated by distances as short as ∼300 km, showed significant differences in allele frequencies indicating barriers to free genetic exchange among areas. Similarly, values for pairwise θ and RST (Table 2) indicated significant overall genetic divergence between most samples. These results suggest significant sub-population structuring on a geographical scale not expected for a non-sessile marine species, and a degree of microgeographic differentiation in S. officinalis around the Iberian Peninsula consistent with the presence of a number of genetic stocks. Whilst this information is relevant to management of the increasing commercial exploitation of this species, additional repeat sampling on a scale of <300 km will be needed to establish if distinct sub-population boundaries are present and temporally stable.
Distinct and significant geographical clines in microsatellite allele frequencies at five out of seven loci were observed extending between the Atlantic and Mediterranean regions sampled within the range of S. officinalis (see Figures 1 and 2). Pérez-Losada et al (1999) also found similar, though less distinct, geographical clines in allozyme allele frequencies between Atlantic and Mediterranean samples of S. officinalis, which they interpreted on the basis of a secondary intergradation model (Endler, 1977). The most commonly proposed causes of clinal patterns in gene frequencies are selection across an environmental gradient, random genetic drift with isolation-by-distance effects, and secondary contact and introgression between previously isolated and genetically divergent populations. Although selection may at least partly explain patterns in genetic variation in several marine species in this area, it is unlikely that this factor can be proposed to account for the clinal patterns observed in the current study: the microsatellite clines are not associated with environmental gradients across the region sampled; there is little evidence for direct selection on microsatellite loci; parallel clines in so many microsatellite and allozyme loci would require improbable levels of genome-wide selection. Mantel tests indicate a significant correlation between geographic and genetic distances, suggesting that the data fit to an isolation-by-distance model of gene flow. Although it is conceivable that isolation-by-distance within the Atlantic-Mediterranean populations of S. officinalis could have created the observed clinal gene frequencies, the presence of parallel clines at so many loci suggest the action of other factors. The connection between the Atlantic Ocean and Mediterranean Sea is known to have been closed in the region of the Strait of Gibraltar several times during glacial periods in the Pliocene and Quaternary (Maldonado, 1985; Bianco, 1990), sometimes associated with substantial environmental changes to the Mediterranean Sea. Although there is no evidence for the historical persistence of S. officinalis in either water body, it is conceivable that this species may have been subdivided into isolated Atlantic and Mediterranean populations during the most recent ice-ages. If current gene flow is restricted, which it appears to be, it is therefore feasible that the present clinal gene frequencies between these areas result from secondary contact and introgression between previously isolated and genetically divergent populations. A possible support for a secondary contact hypothesis is the genotypic disequilibrium observed among three loci in the present study. Inbreeding within isolated populations would produce association between particular alleles at different loci, producing linkage disequilibrium when these populations subsequently intermingle and introgress, the persistence of disequilibrium being dependent on the degree of past inbreeding, the rate of current gene flow and the rate of recombination between loci.
Within all five frequency clines an abrupt change in allele frequencies, supported by highly significant values for allele frequency heterogeneity (exact tests) and genetic divergence (θ and RST values), could be observed between the two SW Mediterranean samples (RMar and Alic) from either side of the Almería-Oran oceanographic front. Genetic discontinuities have been reported in other marine organisms from the same geographical area (Quesada et al, 1995; Sanjuan et al, 1996). Natural selection or restricted gene flow between relatively isolated populations could explain these abrupt genetic changes. As explained above, selection on such a genome-wide scale is unlikely. A hypothesis based on limited gene flow is more plausible: the Almería-Oran front is a strong large-scale ocean current running between Almería (SE Spain) and Oran (Algeria) (Tintore et al, 1988), potentially representing a substantial barrier to migration of coastal cuttlefish. These features suggest that western Mediterranean S. officinalis may be subdivided into two separate gene pools to the SW and NE of the oceanographic front. The front, thus, may represent the contemporary contact point of any Atlantic-Mediterranean introgression, as opposed to the point where physical isolation would have occurred in the past, the Strait of Gibraltar, where there is no evidence of a distinct genetic discontinuity.
Genetic relationships among samples
The unrooted NJ tree (Figure 3) supported the population structure revealed by θ and RST statistics, indicating increasing genetic distance with increasing geographical distance with the most highly supported branchings being between the Atlantic samples and the Mediterranean samples, and between the samples collected from either side of the Almería-Oran front (RMar-Alic/VGel). This tree also supports the suggestion of restricted gene flow within areas representing an Atlantic-Mediterranean division, with the superposition of a contemporary barrier to gene flow at the position of the oceanographic front.
Comparison of microsatellite and allozyme datasets
Allozyme data extracted from Pérez-Losada et al (1999) exhibit a similar degree of global genetic differentiation (θ = 0.062) to the microsatellite data presented here. The scale of pairwise θ is also equivalent in the two data sets, ranging from 0.001 to 0.117 for allozymes (Table 3) and from 0.000 to 0.179 for microsatellites (Table 2). The pattern of relationships among samples however is less clear for allozymes than microsatellites. With allozymes most samples from either side of the Almería-Oran front have consistently high θ between them, but others like SBar (equivalent to Faro) and CPla (equivalent to Alic) do not show significant genetic differences. Allozyme results indicated that θ between geographically proximal samples was not noticeably higher for the pair placed to either side of the oceanographic front (θRMar-CPla = 0.068; CPla = Castellón de la Plana, equivalent to Alic) than for other pairs (eg, θ FFoz-SBar = 0.061; FFoz = Figueira da Foz, equivalent to Lisb; SBar = Sanlúcar de Barrameda, equivalent to Faro). Geographical variation of allele frequencies at polymorphic allozyme loci (see figure 4 of Pérez-Losada et al, 1999) also does not reveal a clear step between RMar and CPla associated to the Almería-Oran oceanographic front. Finally, allozymes also show less geographically concordant relationships among samples: unrooted NJ trees based on D and Dchord gave different topologies [(((GalW,FFoz),RMar),(SBar,VGel))] and less bootstrap support for nodes (72–89%) than NJ trees obtained using microsatellites. These results may indicate that, as found previously in studies of brown trout (Estoup et al, 1998) and squid (Shaw et al, 1999a), microsatellites have greater power than allozymes for resolving genetic relationships among closely related sub-populations of aquatic species with a facility for gene flow on a small geographical scale, and could be more suitable for resolving historical relationships at the intraspecific level.
References
Adcock, GJ, Shaw, PW, Rodhouse, PG, Carvalho, GR (1999). Microsatellite analysis of genetic diversity in the squid Illex argentinus during a period of intensive fishing. Mar Ecol Prog Ser, 187: 171–178.
Bianco, PG (1990). Potential role of the palaeohistory of the Mediterranean and Paratethys basins on the early dispersal of Euro-Mediterranean freshwater fishes. Ichthyol Expl Freshw, 1: 167–184.
Cavalli-Sforza, LL, Edwards, AWF (1967). Phylogenetic analysis: models and estimation procedures. Evolution, 32: 550–570.
Endler, JA (1977). Geographic Variation, Speciation and Clines. Princeton University Press: Princeton, New Jersey.
Estoup, A, Garnery, L, Solignac, M, Cornuet, J -M (1995). Microsatellite variation in the honey bee (Apis mellifera) populations: hierarchical genetic structure and test of the infinite allele and stepwise mutation models. Genetics, 140: 679–695.
Estoup, A, Rousset, F, Michalakis, Y, Cornuet, J -M, Adriamanga, M, Guyomard, R (1998). Comparative analysis of microsatellite and allozyme markers: a case study investigating microgeographic differentiation in brown trout (Salmo trutta). Mol Ecol, 7: 339–353.
Excoffier, L, Smouse, PE, Quattro, JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479–491.
Fisher, RA (1954). Statistical Methods for Research Workers, 12th edn. Oliver and Boyd: Edinburgh.
Goldstein, DB, Linares, AR, Cavalli-Sforza, LL, Feldman, MW (1995). Genetic absolute dating based on microsatellites and the origin of modern humans. Proc Natl Acad Sci USA, 92: 6723–6727.
Goodman, SJ (1997). RST-CALC: A collection of computer programs for calculating unbiased estimates of genetic differentiation and gene flow from microsatellite data and determining their significance. Mol Ecol, 6: 881–885.
Guerra, A (1992). Mollusca, Cephalopoda. In: Ramos MA et al (eds) Fauna Ibérica, vol 1 Museo Nacional de Ciencias Naturales: CSIC, Madrid.
Hughes, CR, Queller, DC (1993). Detection of highly polymorphic microsatellite loci in a species with little allozyme polymorphism. Mol Ecol, 2: 131–137.
Kimura, M, Crow, JF (1964). The number of alleles that can be maintained in a finite population. Genetics, 49: 725–738.
Kimura, M, Ohta, T (1978). Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc Natl Acad Sci USA, 75: 2868–2872.
Maldonado, A (1985). Evolution of the Mediterranean basins and a reconstruction of the Cenozoic palaeoceanography. In: Margalef R (ed) Western Mediterranean, Pergamon Press: London. pp. 18–61.
Naciri, M, Lemaire, C, Borsa, P, Bonhomme, F (1999). Genetic study of Atlantic/Mediterranean transition in sea bass (Dicentrarchus labrax). J Hered, 90: 591–596.
Nei, M (1972). Genetic distance within populations. Am Nat, 106: 283–292.
Nei, M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York.
Parilla, G, Kinder, TH (1992). The physical oceanography of the Alborán Sea. Reports in Meteorol. Oceanography, 40: 143–184.
Pérez-Losada, M (1998). Diferenciación genética y filogenia de Sepia spp. y de otras diez especies de cefalópodos de las familias Ommastrephidae, Loliginidae, Sepiolidae y Octopodidae. PhD Thesis, University of Vigo, Spain.
Pérez-Losada, M, Guerra, A, Sanjuan, A (1999). Allozyme differentiation in the cuttlefish Sepia officinalis (Mollusca: Cephalopoda) from the NE Atlantic and Mediterranean. Heredity, 83: 280–289.
Raymond, M, Rousset, F (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered, 86: 248–249.
Rice, WR (1989). Analyzing tables of statistical test. Evolution, 43: 223–225.
Roldán, MI, García-Marín, JL, Utter, FM, Pla, C (1998). Population genetic structure of European hake, Merluccius merluccius. Heredity, 81: 327–334.
Quesada, H, Zapata, C, Alvarez, G (1995). A multilocus allozyme discontinuity in the mussel Mytilus galloprovincialis: the interaction of ecological and life-history factors. Mar Ecol Prog Ser, 116: 99–115.
Saitou, N, Nei, M (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 4: 406–425.
Sanjuan, A, Comesaña, S, de Carlos, A (1996). Macrogeographic differentiation by mtDNA restriction site analysis in the SW European Mytilus galloprovincialis Lmk. J Exp Mar Biol Ecol, 198: 89–100.
Sanjuan, A, Zapata, C, Alvarez, G (1997). Genetic differentiation in Mytilus galloprovincialis Lmk. throughout the world. Ophelia, 47: 13–31.
Shaw, PW, Pérez-Losada, M (1999). Polymorphic microsatellites in the common cuttlefish Sepia officinalis. Mol Ecol, 9: 237–238.
Shaw, PW, Pierce, GJ, Boyle, PR (1999a). Subtle population structuring within a highly vagile marine invertebrate, the Veneid Squid Loligo forbesi, demonstrated with microsatellite DNA markers. Mol Ecol, 8: 407–417.
Shaw, PW, Turan, C, Wright, JM, O’connell, M, Carvalho, GR (1999b). Microsatellite DNA analysis of population structure in Atlantic herring (Clupea harengus), with direct comparison to allozyme and mtDNA data. Heredity, 83: 490–499.
Takezaki, N, Nei, M (1996). Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics, 144: 389–399.
Tintore, J, La Violette, PE, Blade, I, Cruzado, G (1988). A study of an intense density front in the eastern Alboran sea: the Almería-Oran front. J Phys Oceanogr, 18: 1384–1397.
Weir, BS, Cockerham, CC (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358–1370.
Zane, L, Ostellari, L, Maccatrozzo, L, Bargelloni, L, Cuzin-Roudy, J, Buchholz, F, Patarnello, T (2000). Genetic differentiation in a pelagic crustacean (Meganyctiphanes norvegica: Euphausiacea) from the North East Atlantic and the Mediterranean Sea. Mar Biol, 136: 191–199.
Acknowledgements
We are grateful to A Vila and JL Sánchez for assistance in the collection of the samples. Thanks are also due to A Merino for her valuable help. This research was partially supported by project AMB94-0371 (CICYT, Spain). MP-L was supported by a fellowship from University of Vigo (Spain).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pérez-Losada, M., Guerra, A., Carvalho, G. et al. Extensive population subdivision of the cuttlefish Sepia officinalis (Mollusca: Cephalopoda) around the Iberian Peninsula indicated by microsatellite DNA variation. Heredity 89, 417–424 (2002). https://doi.org/10.1038/sj.hdy.6800160
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800160
Keywords
This article is cited by
Interannual fluctuations in connectivity among crab populations (Liocarcinus depurator) along the Atlantic-Mediterranean transition
Scientific Reports (2022)
Genetic Differentiation Among Populations of Octopus minor Based on Simple Sequence Repeats Mined from Transcriptome Data
Journal of Ocean University of China (2022)
Morphological characteristics and genetic differentiation of Lutraria maxima in coast waters off southeast China
Journal of Oceanology and Limnology (2021)
Genetic diversity and population structure of the rockpool shrimp Palaemon elegans based on microsatellites: evidence for a cryptic species and differentiation across the Atlantic–Mediterranean transition
Scientific Reports (2020)
Genetic population structure and phylogeny of the common octopus Octopus vulgaris Cuvier, 1797 in the western Mediterranean Sea through nuclear and mitochondrial markers
Hydrobiologia (2018)