Porcine Models of the Intestinal Microbiota: The Translational Key to Understanding How Gut Commensals Contribute to Gastrointestinal Disease
- Department of Clinical Sciences, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
In the United States, gastrointestinal disorders account for in excess of $130 billion in healthcare expenditures and 22 million hospitalizations annually. Many of these disorders, including necrotizing enterocolitis of infants, obesity, diarrhea, and inflammatory bowel disease, are associated with disturbances in the gastrointestinal microbial composition and metabolic activity. To further elucidate the pathogenesis of these disease syndromes as well as uncover novel therapies and preventative measures, gastrointestinal researchers should consider the pig as a powerful, translational model of the gastrointestinal microbiota. This is because pigs and humans share striking similarities in their intestinal microbiota as well as gastrointestinal anatomy and physiology. The introduction of gnotobiotic pigs, particularly human-microbial associated pigs, has already amplified our understanding of many gastrointestinal diseases that have detrimental effects on human health worldwide. Continued utilization of these models will undoubtedly inform translational advancements in future gastrointestinal research and potential therapeutics.
The Rise and Plateau of Gut Microbiota Research
Over the first few days of life, the neonatal gut is rapidly populated by a diverse population of microorganisms that exponentially expands to a number exceeding that of the total host cells (1). Interestingly, a recently-proposed paradigm shift suggests that the gut microbiota begins to develop in utero rather than during birth (2–5). Such findings are pivotal to our understanding of the gut microbiota given that microbes support vital physiologic processes including production of volatile fatty acids and vitamin K as well as transportation of electrolytes and water across the mucosal surface. The presence of gut commensals is particularly crucial in developing neonates, the developing immune system of which is stimulated by indigenous microflora. Furthermore, the absence of certain gut commensals has been associated with increased risk for lifelong autoimmune diseases (6, 7).
Paralleling increased interest in the gut bacterial microbiota, herein referred to as the gut microbiota, there have been robust advancements in laboratory technologies able to characterize such populations. Increased accessibility to 16S rRNA and next-generation DNA sequencing has permitted detailed characterization of the gastrointestinal microbiota in numerous species. Furthermore, “-omics” laboratory techniques have informed investigations into the systemic impact of gut microbial communities. These communities are now heavily linked with the development of several intestinal and multisystemic diseases in humans including obesity and inflammatory bowel disease (IBD).
Despite instrumental advances in microbial identification, establishing association between variations in microbial diversity and disease phenotypes is continuously challenged by sampling limitations within humans. Subsequently, gastrointestinal research has experienced drastic drops in the development of new pharmaceuticals and diagnostics despite large monetary investments by public and private institutions. This decelerated translation of basic research to clinical practice is colloquially referred to as the “pipeline problem” (8). This problem is attributable, at least in part, to the historic use of inappropriate animal models, frequently rodents, that do not adequately emulate the human patient. Consequently, gastrointestinal research has steadily turned toward pig models given their anatomic and physiologic similarities with humans (9). This review examines these similarities and summarizes recently-characterized parallels between the pig and human gut microbiota, thereby advocating for the pig as an unequaled model of the human gut microbiota and a powerful tool to break through current plateaus in exploring disease association with the gut microbiota.
Anatomic and Physiological Similarities Between the Human and Pig Outweigh Differences
Nutritionists around the world continue to preach that we are what we eat. In the realm of gastroenterology, this proverb can be interpreted literally given that gastrointestinal morphology is directly associated with meal frequency, type of food from which nutrients are extracted and composition of the gut microbiota (10, 11). Unlike rats and other domestic animals, the pig and human are both true omnivores (12). Therefore, it comes as no surprise that their gastrointestinal tracts share many macroscopic and microscopic features (Figure 1).
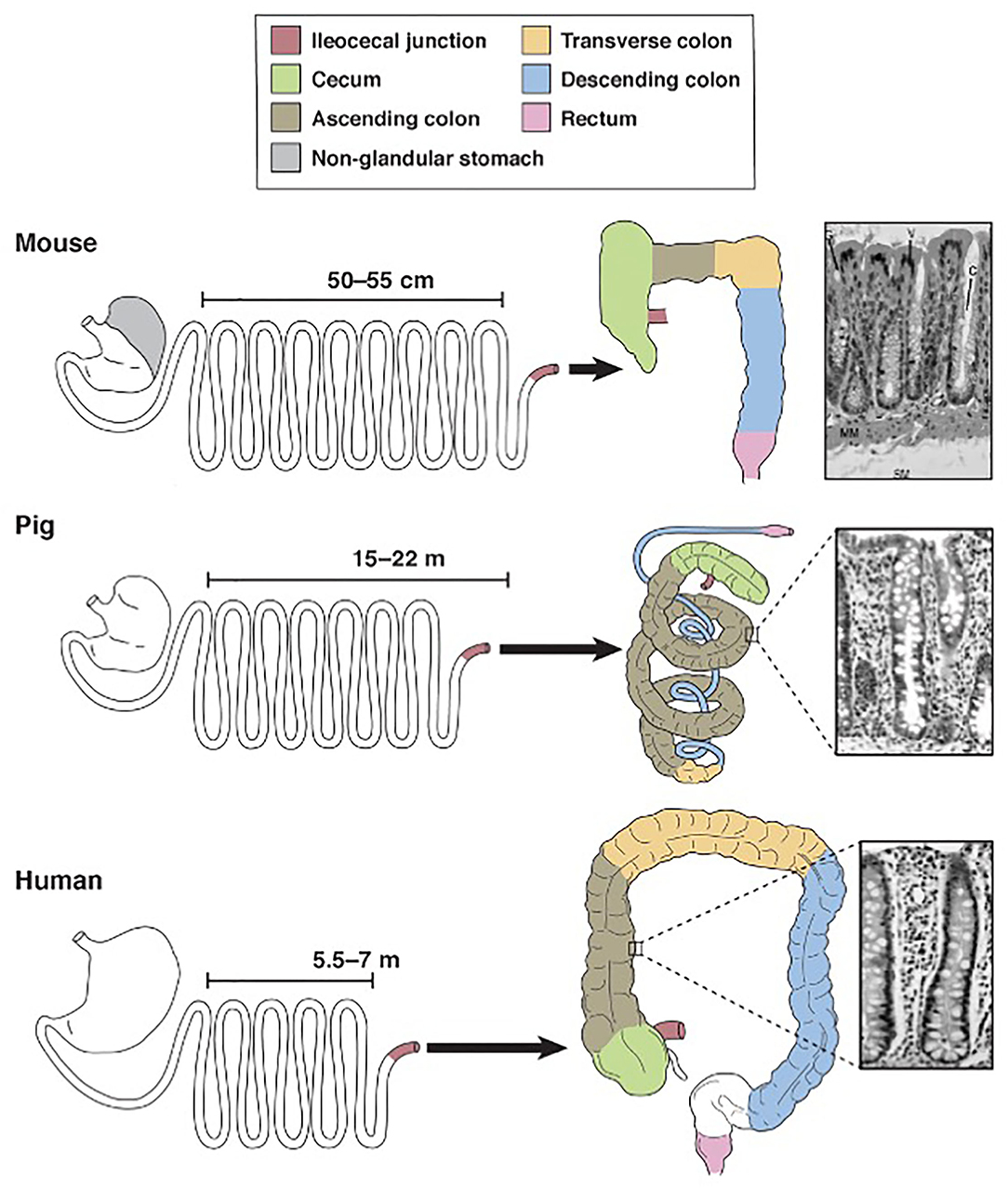
Figure 1. Schematic diagram for comparison of mouse, pig and human gastrointestinal tract anatomy. Derived from Ziegler et al. (13).
Pigs, rodents and humans all have a “simple” stomach comprised of one compartment. While the entirety of the human stomach and majority of the pig stomach are glandular, the rodent stomach is divided into a glandular portion and non-glandular portion. This non-glandular portion, which is used for food storage and digestion, defines nearly 50% of the gastric mucosal surface area in rodents, therefore complicating the use of rodent models for human gastric studies. In all three species, the glandular section is composed of cardiac, gastric and pyloric mucosa. The pig stomach contains significantly more cardiac mucosa than the human stomach. This cardiac mucosa creates a pseudo-diverticulum for food storage as well as digestion and is a proposed site of microbial metabolism. Furthermore, cardiac epithelial cells are mainly mucus-secreting while parietal and chief cells of the gastric and pyloric mucosa secrete hydrochloric acid and pepsinogen, respectively. Taking these physiochemical differences into account, the relatively large size of the cardiac mucosa in pigs may support a unique, physiologic niche for microbes that is not mirrored in the human. Therefore, caution is urged when making comparisons between human and pig gastric microbiota. Researchers should be aware of this anatomic variation and consider sampling protocols that emphasize collection from the shared gastric and pyloric mucosa rather than the cardiac mucosa.
Aboard to the stomach, porcine and human intestines are strikingly similar. The ratio of total intestinal length per kilogram bodyweight is ~ 0.1 in both pigs and humans, meaning both species share a similar relative length of their intestinal tract (14, 15). The small intestines of both species are macroscopically similar, characterized by a linear, continuous tube anchored to the peritoneum by intricately-vascularized mesentery. This linear morphology is retained in the human large intestines but disrupted in the pig by the formation of a spiral colon that coils into itself. The pig colon is additionally slightly larger than the human colon; the pig colon accounts for approximately 46% of total intestinal tract weight while the human colon accounts for approximately 36% of total intestinal tract weight (14). Despite this discrepancy in shape and relative size, however, the large intestines in both species are functionally similar and house the body's largest population of microbes. Notably, the large intestinal microbiota in both species is responsible for synthesizing volatile fatty acids, which are absorbed through the intestinal mucosa (16). Sacculations and tenia within the human and pig colon additionally provide similar physiologic and anatomic niches for gut commensals and thereby may encourage similar microbial populations.
The colon is also the primary site for ingesta fermentation in both pigs and humans. Rodents, however, are cecal fermenters. Given that intestinal fermentation is primarily regulated by luminal microbes, its anatomic localization directly affects the constituents of the gut microbiota. Therefore, given that rodents ferment within their cecum, the constituents of their large intestinal microbiota will significantly vary from that of colon fermenters, namely pigs and humans. The pig and human ceca do diverge with respect to their size; the pig cecum is relatively large and grossly demarcated from the remainder of the large intestines compared to the human cecum. Therefore, similar to the gastric cardia, the study of the pig cecum as a model for humans must be regarded with caution.
Mucosal Peyer's patches are another anatomic feature that distinguishes the pig and human gastrointestinal tract, specifically the small intestines, from one another. Pig and human Peyer's patches intestines diverge with respect to the cellular composition, development, distribution and number (12, 17, 18). In fact, organogenesis research suggests that pigs develop two distinct types of Peyer's patches, jejunal and ileal, while humans develop only one (19). Given that one of the Peyer's patches many functions is to discriminate between pathogenic and commensal bacteria, we can speculate that this interspecies variability may provoke distinct host perception of and interaction with gut commensals. Conversely, several researchers argue that these differences between pig and human Peyer's patches are of limited significance because pigs and humans demonstrate similar immunologic responses to various gastrointestinal insults (12, 20, 21).
The anatomic similarities between the pig and human gastrointestinal tracts translate to comparable intestinal motility, referring to the contraction and relaxation of the muscularis layers, as well as similar ingesta transit time, referring to the time ingesta takes to travel from the esophagus to the rectum (22). Significant alterations in the gut microbiota are regularly detected in individuals with altered intestinal mobility, such as surgery patients with postoperative ileus. Therefore, similar ingesta transit times between pigs and humans may facilitate similar populations of gut microbiota by discouraging colonization by pathogenic or non-commensal organisms.
Comparing the Neonatal and Adult Intestinal Microbiota of Pigs and Humans Reveals Important Similarities and Differences
Many studies have made impactful comparisons between the intestinal microbiota of pigs and humans (23–25). Although the intestinal microbiota in healthy adults and mature pigs is relatively stable, it fluctuates widely over the first year of life for both species (26–28). One study has demonstrated that piglets from the same litter as well as newborn human twins can differ with respect to their intestinal microbiota (29). Furthermore, adult humans and pigs demonstrate similar alterations to their gut microbiota in response to environmental stressors and antibiotics (30–35). This suggests that the pig is a powerful model of pathologic disturbances in the intestinal microbiota, such as those elicited by antibiotic-induced dysbiosis and IBD.
Notably, most studies on the intestinal microbiota are based primarily on fecal samples due to limitations in sample acquisition. Sampling the human gastrointestinal microbiota is particularly challenging given that elective surgeries to obtain such samples are costly, time-consuming, and simply unappealing to most individuals. Elective surgeries in laboratory animals are certainly more feasible but still elicit a systemic stress response, which may compromise the microbiota and consequently the study's integrity. Therefore, very few studies have provided direct, interspecies comparisons of microbial populations across specific sites of the gastrointestinal tract.
Given this sampling limitation, most comparative studies have been limited to the large intestines, namely the colon (Figure 2). Under natural conditions, more than 90% of the bacteria in the colon of adult humans and pigs are within one of two phyla: Firmicutes or Bacteroidetes (27, 30, 31, 33, 34, 38). Although there is slight variation in bacterial genus and species due to species specificity, shared bacterial physiology and metabolism within these phyla solidify the adult pig as a feasible model of the human colonic microbiota in health.
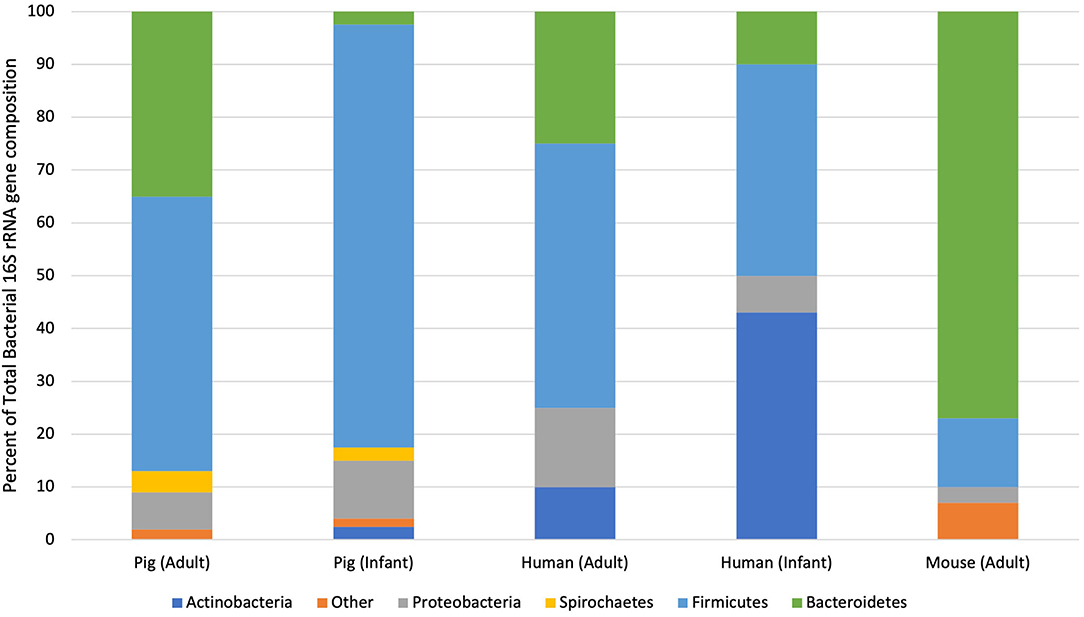
Figure 2. Taxonomic distribution of bacterial phyla from pigs, humans and mice at different life stages. This graph illustrates the percent of sequences assigned to each bacterial phylum isolated through 16S rRNA sequencing of pooled fecal samples isolated from healthy individuals of each species (33, 36, 37).
Species divergence becomes more readily apparent when considering the large intestinal microbiota of neonatal pigs and human infants. Large intestinal content and feces isolated from adults and infants contain significantly more Actinobacteria than adult or neonatal pigs (33, 39). In fact, the large intestines of formula- and breast-fed infants are dominated by Actinobacteria. This discrepancy between wildtype neonatal pigs and human infants can be mitigated through the use of increasingly-available, gnotobiotic animals, which will be further discussed in subsequent sections. Wildtype piglets may remain relevant models of the infant gut microbiota, however, given their shared alternations in the intestinal microbiota following the introduction of solid food (40). Therefore, wildtype neonatal pigs can be used to examine the pathogenesis and potential therapies for dietary and environmental perturbations on the intestinal microbiota of infants as long as relative alterations in specific bacterial genera and species are interpreted with care.
Although the large intestines of adults and mature pigs are dominated by the same two phyla, interspecies variability is further elucidated by comparing the bacterial genera isolated from those phyla. Within Bacteroidetes, the most abundant genus in the human intestines is Bacteroides while that in pig intestines is Prevotella (26). At 10 weeks-of-age, Prevotella represents up to 30% of the microbiota of the pig colon. As the pig reaches 22-weeks-old, however, relative numbers of Prevotella species drop to 4% and relative numbers of Anaerobacter sp., which are in the Firmicutes phylum, increase (33, 41). This steady decline in Bacteroidetes and increase in Firmicutes is mirrored in human infants over their first 4 months of life (41, 42). Therefore, although there are select differences in bacterial genera and species between pig and human intestines, shared bacterial phyla likely trigger comparable physiologic developments and establish similar symbiotic relationships.
One of the most conspicuous dissimilarities between the human and pig microbiota is the presence of specific microorganisms that are unique to pigs. In fact, both sow-reared and formula-fed piglets have greater intestinal microbial diversity than human infants (39). Low numbers of Fusobacterium are detected in the feces of neonatal pigs but not breast-fed infants (39). Significantly more Lactobacilli, Spirochetes and Streptococci are isolated from the porcine intestines than the human intestines at any age (26, 33, 41). A high percentage of the porcine ileal microbiota is represented by Proteobacterium, which is not reported in the human ileum (30). However, large numbers of Proteobacteria have be isolated from the feces of breast-fed infants (39). This being said, aforementioned limitations on sample collection presumably limit researchers' ability to fully characterize every bacterium within the human intestines. Identification of certain gastrointestinal commensals in pigs but not humans may be attributable to sampling techniques that are feasible in the former but not the latter.
The stark differences among the environments of humans, pigs and rodents must also be considered as a potential driver of these variations. Coprophagy is considered to be normal behavior of pigs and rodents. In piglets, the influence of sanitary conditions and coprophagia on the gastrointestinal microbiota has been well established (43, 44). Coprophagia of the sow's feces may in fact benefit piglets as a source for commensal microbes that foster the development of the piglet's microbiome. Considering the commonality of coprophagia in pigs, many researchers have turned to germ free, gnotobiotic or even humanized pigs, all of which are further discussed below.
Despite select dissimilarities in the intestinal microbiota of pigs and humans, the alluring nature of the pig as a model for the human intestinal microbiota is largely attributable to the fact that the pig is the best model we currently have. While the laboratory animal population across the world remains dominated by rodents, anatomic and physiologic dissimilarities between the human and rodent gastrointestinal tracts promote significant interspecies variation in the gut microbiota (23, 45). Nearly 85% of the bacteria genera isolated from the mouse gut is not present within the human, which immensely overshadows aforementioned variations in intestinal microbiota of pigs and humans (46).
Cutting-Edge Techniques are Available in Pig Models
The pig's potential for high impact, translational research is further exemplified by the vast number of laboratory techniques that have been adapted to the pig. One of the biggest advantages of pig is its relative size compared to other traditional laboratory animals such as mice and rats. The pig's large gastrointestinal tract equates to increased surgical access and manipulation as well as experimental tissue volume. Furthermore, standardization of pig care and surgery permits even a novice researcher to easily expand their studies to include pig models (47–50).
Paralleling this expanse in laboratory techniques, there has been a sharp rise in the type of available pig models. The pig genome has been fully sequenced, leading to the emergence of genetically-modified pigs. Genetically-modified pigs are stronger translational models than rodents given that the human genome is more closely related to the pig than to the mouse or rat (51–53). Such models can strengthen our understanding of disease pathogenesis by introducing the ability to knock-down or knock-out genes as well as artificially tag specific cell populations and proteins so that they can be traced along the course of disease. Therefore, observations elucidated from these pigs can better inform putative disease therapies in human medicine.
In addition to genetically-modified pigs, many researchers are introducing germ-free, gnotobiotic and human-microbial associated (HMA) pigs to their experimental design. Through elimination of the intestinal microbiota in germ-free pigs, investigators may track variances in disease pathogenesis and consequently infer disease association or correlation with specific gut commensals. This being said, germ-free pigs have shorter small intestines, shorter crypts, longer villi and smaller Peyer's patches compared to conventionally-raised pigs (39). These anatomic variances, along with associated alterations in intestinal physiology and immunology, force researchers to question the translatability of germ-free pigs. Gnotobiotic pigs are arguably more powerful models of gastrointestinal disease because they permit direct manipulation of the intestinal microbiota such that investigators can characterize disease progression in the presence of select microbes. One emerging subset of gnotobiotic pigs, HMA pigs, are exceptionally noteworthy and will be discussed further within the following section.
Important Differences Between the Human and Porcine Intestinal Microbiota May Be Surpassed Through Human-Microbial Associated Pigs
Several laboratories have successfully “humanized” the intestinal microbiota of animal models by inoculating germ-free animals with microorganisms isolated from the human intestines (11). Human microbiota-associated piglets have been established using inocula from infants, children and adults (40, 54, 55). The gut microbiota from these recipient HMA piglets is more similar to that of the human donor than that of conventionally-raised piglets. Furthermore, age-related microbial succession in HMA piglets mirrors that observed within the human donors (40).
Notably, bacteria from the Actinobacteria phyla, namely Bifidobacteria, successfully colonize HMA piglets and reach population densities similar to those in humans (40). Therefore, although the intestinal microbiota of conventionally-raised piglets diverges from that of infants due to the absence of naturally occurring Acintobacteria, the intestinal microbiota of HMA-piglets can be manipulated such that it closely emulates that of humans. Attempts to create HMA models in other animal species have not been as fruitful. The predominant bacterial genera of the human intestines, including Lactobacillus and Bifidobacterium, do not successfully colonize the gut of mice and zebrafish following inoculation (56–58). Therefore, not all HMA animals are powerful models of the human intestinal microbiota and something within the pig, perhaps the similarities in gastrointestinal anatomy and physiology, supports successful colonization of human bacteria.
The strength in the HMA pig model stems from the vast number of questions that can be gleaned through its integration in experimental design. As probiotics and prebiotics continue to gain momentum while synbiotics and postbiotics gain traction in the world of dietary supplementation, HMA pig models may be key to evaluating the impact of these compounds on the microbiota (59–61). Long-term studies of HMA pigs can therefore overcome the limitations in sample collection from the intestinal lumen of humans, thereby allowing further study of microbial community succession and biogeography in infants to adults. Additionally, it will be interesting to use HMA pigs to investigate whether key microbial activities can be transferred to recipient animals. This may inform future treatment modalities for gastrointestinal disorders that are associated with microbiota derangements.
With Impactful Models Comes Impactful Costs
One of the biggest limitations of pig models is their relative cost of model acquisition and maintenance. In the Unites States, the per diem housing cost for rodents typically ranges between $1-3 USD while for a pig is up to $19 USD depending on the facility1. Given that the lifespan of most laboratory rodents is around 2–3 years and that of pigs is around 20 years, accruing costs of pig colony management may limit the feasibility of long-term, prospective studies. Even a single finishing pig, which is between 4-months and 1-year-old, is up to 30 times the cost of an adult laboratory mouse or rat (personal communication with Dr. Jack Odle, North Carolina State University). Despite the previously discussed power of gnotobiotic and HMA pig models, the limited number of laboratory facilities that can house large animals significantly potentiates costs. One litter of gnotobiotic pigs is estimated to cost around $25,000 and the cost of a 9-month study on 2 gnotobiotic litters is around $350,000 (personal communication with Dr. Michael Oglesbee, The Ohio State University). While gnotobiotic mice are certainly not cheap at $500 each, they are significantly less expensive than their porcine counterpart2.
Concluding Remarks and Future Directions
As gastrointestinal researchers continue to embrace the pig model and HMA pigs become increasingly accessible, emerging translational studies will divulge the pathogenesis of and putative therapies for diseases characterized by or associated with intestinal dysbiosis. Pig models have already proven advantageous in the study of microbiota-associated diseases such as necrotizing enterocolitis of infants, which is a complication of preterm, very-low birth weight infants that has been associated with bacterial colonization of the intestines (62–70). Particularly now that the porcine intestinal microbiota has been fully characterized through multiple life stages, gastrointestinal researchers can easily track changes in the microbial composition and make direct associations between these changes and disease (28, 38, 41, 71). Compounding these studies with investigations into potential preventative and therapeutic interventions will undoubtedly uncover exciting, translational advances that will benefit humans and animals alike.
Author Contributions
ER: manuscript generation. AB and AZ: editing and revising. All authors contributed to the article and approved the submitted version.
Funding
ER was supported by a COHA Translational Fellowship funded by U01 TR002953.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^NCSU CVM Laboratory Animal Resources. https://cvm.ncsu.edu/lar-cost/
2. ^Taconic Biosciences Inc. https://www.taconic.com/mouse-model/black-6-b6ntac
References
1. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutalism in the human intestine. Science. (2005) 307:1104816. doi: 10.1126/science.1104816
2. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. (2016) 6:23129. doi: 10.1038/srep23129
3. Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS ONE. (2015) 10:e0133320. doi: 10.1371/journal.pone.0133320
4. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. (2014) 6, 237ra265. doi: 10.1126/scitranslmed.3008599
5. Stinson LF, Boyce MC, Payne MS, Keelan JA. The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front Microbiol. (2019) 10:1124. doi: 10.3389/fmicb.2019.01124
6. Savage DC. Microbial ecology of the gastrointestinal tract. Ann Rev Microbiol. (1977) 31:543. doi: 10.1146/annurev.mi.31.100177.000543
7. Leslie M. Gut microbes keep rare immune cells in line. Science. (2012) 335:1428. doi: 10.1126/science.335.6075.1428
8. Phillips KVBS, Issa A. Diagnostics and biomarker development: priming the pipeline. Nat Rev Drug Discov. (2006) 5:nrd2033. doi: 10.1038/nrd2033
9. Lunney JK, Van Goor A, Walker KE, Hailstock T, Franklin J, Dai C. Importance of the pig as a human biomedical model. Sci Transl Med. (2021) 13:abd5758. doi: 10.1126/scitranslmed.abd5758
10. Foong JPP, Hung LY, Poon S, Savidge TC, Bornstein JC. Early life interaction between the microbiota and the enteric nervous system. Am J Physiol Gastrointest Liver Physiol. (2020) 319:G541–8. doi: 10.1152/ajpgi.00288.2020
11. Yu Y, Lu L, Sun J, Petrof EO, Claud EC. Preterm infant gut microbiota affects intestinal epithelial development in a humanized microbiome gnotobiotic mouse model. Am J Physiol Gastrointest Liver Physiol. (2016) 311:G521–32. doi: 10.1152/ajpgi.00022.2016
12. Kararli TT. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos. (1995) 16:351–80. doi: 10.1002/bdd.2510160502
13. Ziegler A, Gonzalez L, Blikslager A. Large animal models: the key to translational discovery in digestive disease research. Cell Mol Gastroenterol Hepatol. (2016) 2:6. doi: 10.1016/j.jcmgh.2016.09.003
14. Gonzalez LM, Moeser AJ, Blikslager AT. Porcine models of digestive disease: the future of large animal translational research. Transl Res. (2015) 166:12–27. doi: 10.1016/j.trsl.2015.01.004
15. Hatton GB, Yadav V, Basit AW, Merchant HA. Animal farm: considerations in animal gastrointestinal physiology and relevance to drug delivery in humans. J Pharm Sci. (2015) 104:2747–76. doi: 10.1002/jps.24365
16. Stevens C. Physiologicla implications of microbial digestion in the large intestine of mammals: relation to dietary factors. Am J Clin Nutr. (1978) 10:S161–8. doi: 10.1093/ajcn/31.10.S161
17. Maroilley T, Berri M, Lemonnier G, Esquerre D, Chevaleyre C, Melo S, et al. Immunome differences between porcine ileal and jejunal Peyer's patches revealed by global transcriptome sequencing of gut-associated lymphoid tissues. Sci Rep. (2018) 8:9077. doi: 10.1038/s41598-018-27019-7
18. Burkey TE, Skjolaas KA, Minton JE. Board-invited review: porcine mucosal immunity of the gastrointestinal tract. J Anim Sci. (2009) 87:1493–501. doi: 10.2527/jas.2008-1330
19. Furukawa M, Ito S, Suzuki S, Fuchimoto D, Onishi A, Niimi K, et al. Organogenesis of ileal Peyer's patches is initiated prenatally and accelerated postnatally with comprehensive proliferation of B cells in pigs. Front Immunol. (2020) 11:604674. doi: 10.3389/fimmu.2020.604674
20. Swindle MM, Makin A, Herron AJ, Clubb FJ, Frazier KS. Swine as models in biomedical research and toxicology testing. Vet Pathol. (2012) 49:344–56. doi: 10.1177/0300985811402846
21. Swindle MM. Comparative anatomy and physiology of the pig. Scand J Lab Anim Sci. (1998) 25:11–21.
22. Graham H, Aman P. The pig as a model in dietary fibre digestion studies. Scand J Gastroenterol Suppl. (1987) 129:55–61. doi: 10.3109/00365528709095851
23. Li X, Liang S, Xia Z, Qu J, Liu H, Liu C, et al. Establishment of a Macaca fascicularis gut microbiome gene catalog and comparison with the human, pig, and mouse gut microbiomes. Gigascience. (2018) 7. doi: 10.1093/gigascience/giy100
24. Xiao L, Estelle J, Killerich P, Ramayo-Caldas Y, Zia Z, Feng Q, et al. A reference gene catalogue of the pig gut microbiome. Nat Microbiol. (2016) 1:16161. doi: 10.1038/nmicrobiol.2016.161
25. Chen C, Zhou Y, Fu H, Xiogn X, Fang S, Jiang H, et al. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat Commun. (2021) 12. doi: 10.1038/s41467-021-21295-0
26. Roura E, Koopmans SJ, Lalles JP, Le Huerous-Luron I, de Jager N, Schuurman T, et al. Critical review evaluating the pig as a model for human nutritional physiology. Nutr Res Rev. (2016) 29:60–90. doi: 10.1017/S0954422416000020
27. Gao S, Yan L, Wang R, Li J, Yong J, Zhou X, et al. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat Cell Biol. (2018) 20:721–34. doi: 10.1038/s41556-018-0105-4
28. Chen L, Xu Y, Chen X, Fang C, Zhao L, Chen F. The maturing development of gut microbiota in commercial piglets during the weaning transition. Front Microbiol. (2017) 8:1688. doi: 10.3389/fmicb.2017.01688
29. Koo H, Hakim JA, Crossman DK, Lefkowitz EJ, Morrow CD. Sharing of gut microbial strains between selected individual sets of twins cohabitating for decades. PLoS ONE. (2019) 14:e0226111. doi: 10.1371/journal.pone.0226111
30. Isaacson R, Kim HB. The intestinal microbiome of the pig. Anim Health Res Rev. (2012) 13:100–9. doi: 10.1017/S1466252312000084
31. Looft T, Johnson A, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Nat Acad Sci. (2012) 109:1691–6. doi: 10.1073/pnas.1120238109
32. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. (2008) 6:e280. doi: 10.1371/journal.pbio.0060280
33. Lamendella R, Santo Domingo JW, Ghosh S, Martinson J, Oerther DB. Comparative fecal metagenomics unveils unique functional capacity of the swine gut. BMC Microbiol. (2011) 11:103. doi: 10.1186/1471-2180-11-103
34. Guo X, Xia X, Tang R, Zhou J, Zhao H, Wang K, et al. Development of a real-time PCR method for Firmicutes and Bacteroidetesin faeces and its application to quantify intestinal population of obese and lean pigs. Lett Appl Microbiol. (2008) 47:367–73. doi: 10.1111/j.1472-765X.2008.02408.x
35. Borewicz KA, Kim HB, Singer RS, Gabhart CJ, Sreevatsan S, Johnson T, et al. Changes in the porcine intestinal microbiome in response to infection with Salmonella enterica and Lawsonia intracellularis. PLoS ONE. (2015) 10:e0139106. doi: 10.1371/journal.pone.0139106
36. Schokker D, Zhang J, Zhang L-L, Vastenhouw HG, Heilig HJ, Smidt H, et al. Early-life environmental variation affects intestinal microbiota and immune development in new-born piglets. PLoS ONE. (2014) 9:e100040. doi: 10.1371/journal.pone.0100040
37. Yang Y-W, Chen M-K, Yang B-Y, Huang X-J, Zhang X-R, He L-Q, et al. Use of 16S rRNA gene-targeted group-specific primers for real-time pcr analysis of predominant bacteria in mouse feces. Appl Environ Microbiol. (2015) 81:6749–56. doi: 10.1128/AEM.01906-15
38. Crespo-Piazuelo D, Migura-Garcia L, Estelle J, Criado-Mesas L, Revilla M, Castello A, et al. Association between the pig genome and its gut microbiota composition. Sci Rep. (2019) 9:8791. doi: 10.1038/s41598-019-45066-6
39. Wang M, Donovan SM. Human microbiota-associated swine: current progress and future opportunities. ILAR J. (2015) 56:63–73. doi: 10.1093/ilar/ilv006
40. Pang X, Hua X, Yang Q, Ding D, Che C, Cui L, et al. Inter-species transplantation of gut microbiota from human to pigs. ISME J. (2007) 1:156–62. doi: 10.1038/ismej.2007.23
41. Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, et al. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet Microbiol. (2011) 153:124–33. doi: 10.1016/j.vetmic.2011.05.021
42. Vaiserman A, Romanenko M, Piven L, Moseiko V, Lushchak O, Kryzhanovska N, et al. Differences in the gut firmicutes to bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. (2020) 20:1–8. doi: 10.1186/s12866-020-01903-7
43. Montagne L, Arturo-Schaan M, Le Floc'h N, Guerra L, Le Gall M. Effect of sanitary conditions and dietary fibre on the adaptation of gut microbiota after weaning. Livestock Sci. (2010) 133:113–6. doi: 10.1016/j.livsci.2010.06.039
44. Schmidt B, Mulder IE, Musk CC, Aminov RI, Lewis M, Stokes CR, et al. Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS ONE. (2011) 6:e28284. doi: 10.1371/journal.pone.0028284
45. Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, et al. A catalog of the mouse gut metagenome. Nat Biotechnol. (2015) 33:1103–8. doi: 10.1038/nbt.3353
46. Nguyen TLA, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Disease Models Mechanisms. (2015) 8:1–16. doi: 10.1242/dmm.017400
47. Rehbinder C, Baneux P, Forbes D, Van Herck H, Nicklas W, Rugaya Z, et al. FELASA recommendations for the health monitoring of breeding colonies and experimental units of cats, dogs and pigs: Report of the Federation of European Laboratory Animal Science Associations (FELASA) Working Group on Animal Health. Lab Anim. (1998) 32:1–17. doi: 10.1258/002367798780559428
48. Marchant-Forde JN, Herskin MS. Pigs as laboratory animals. In: Advances in Pig Welfare. WoodHead Publishing (2018). p. 445–75.
49. Smith AC, Swindle MM. Preparation of swine for the laboratory. ILAR J. (2006) 47:358–63. doi: 10.1093/ilar.47.4.358
50. Swindle M. Swine in the Laboratory. Surgery, Anaesthesia, Imaging and Experimental Techniques. Boca Raton: CRC Press/Taylor and Friends (2007).
51. Wernersson R, Schierup MH, Jorgensen FG, Gorodkin J, Panitz F, Staerfeldt H-H, et al. Pigs in sequence space: A 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genom. (2005) 6:1–7. doi: 10.1186/1471-2164-6-70
52. Jørgensen F, Hobolth A, Hornshoj H, Bendizen C, Fredhold M, Schierup M. Comparative analysis of protein coding sequences from human, mouse and the domestic pig. BMC Biol. (2005) 3:2. doi: 10.1186/1741-7007-3-2
53. Walters EM, Wolf E, Whyte JJ, Mao J, Renner S, Nagashima H, et al. Completion of the swine genome will simplify the production of swine as a large animal biomedical model. BMC Med Genomics. (2012) 5:55. doi: 10.1186/1755-8794-5-55
54. Zhang H, Wang H, Shepherd M, Wen K, Li G, Yang X, et al. Probiotics and virulent human rotavirus modulate the transplanted human gut microbiota in gnotobiotic pigs. Gut Pathog. (2014) 6:1–7. doi: 10.1186/s13099-014-0039-8
55. Zhang Q, Widmer G, Tzipori S. A pig model of the human gastrointestinal tract. Gut Microbes. (2013) 4:193–200. doi: 10.4161/gmic.23867
56. Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. (2006) 127:423–33. doi: 10.1016/j.cell.2006.08.043
57. Imaoka A, Setoyama H, Takagi A, Matsumoto S, Umesaki Y. Improvement of human faecal flora-associated mouse model for evaluation of the functional foods. J Appl Microbiol. (2004) 96:656–63. doi: 10.1111/j.1365-2672.2004.02189.x
58. Raibaud P, Ducluzeau R, Dubos F, Hudault S, Bewa H, Muller MC. Implantation of bacteria from the digestive tract of man and various animals into gnotobiotic mice. Am J Clin Nutr. (1980) 33:2440. doi: 10.1093/ajcn/33.11.2440
59. Scarpellini E, Rinninella E, Basillico M, Colomier E, Rasetti C, Larussa T, et al. From pre- and probiotics to post-biotics: a narrative review. Int J Environ Res Public Health. (2021) 19:37. doi: 10.3390/ijerph19010037
60. Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, et al. The international scientific association of probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. (2021) 18:649–67. doi: 10.1038/s41575-021-00440-6
61. Li H-Y, Zhou D-D, Gan R-Y, Huang S-Y, Zhao C-N, Shang A, et al. Effects and mechanisms of probiotics, prebiotics, synbiotics, and postbiotics on metabolic diseases targeting gut microbiota: a narrative review. Nutrients. (2021) 13:3211. doi: 10.3390/nu13093211
62. Bjornvad CR, Thymann T, Deutz NE, Burrin DG, Jensen SK, Jensen BB, et al. Enteral feeding induces diet-dependent mucosal dysfunction, bacterial proliferation, and necrotizing enterocolitis in preterm pigs on parenteral nutrition. Am J Physiol Gastrointest Liver Physiol. (2008) 295:G1092–1103. doi: 10.1152/ajpgi.00414.2007
63. Sangild PT, Siggers RH, Schmidt M, Elnif J, Bjornvad CR, Thymann T, et al. Diet- and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterology. (2006) 130:1776–92. doi: 10.1053/j.gastro.2006.02.026
64. Mendez YS, Khan FA, Perrier GV, Radulescu A. Animal models of necrotizing enterocolitis. World J. Pediatric Surg. (2020) 3:e000109. doi: 10.1136/wjps-2020-000109
65. Fitzgibbons SC, Ching Y, Yu D, Carpenter J, Kenny M, Weldon C, et al. Mortality of necrotizing enterocolitis expressed by birth weight categories. J Pediatr Surg. (2009) 44:1072–6. doi: 10.1016/j.jpedsurg.2009.02.013
66. Krediet T, Lelyveld N, Vijibrief D, Brouwers H, Kramer W, Fleer A, et al. (2007). Microbiological factors associated with neonatal necrotizing enterocolitis: protective effect of early antibiotic treatment. Acta Paediatr. 92:1180–2. doi: 10.1111/j.1651-2227.2003.tb02481.x
67. Ladd N, Ngo T. The use of probiotics in the prevention of necrotizing enterocolitis in preterm infants. Baylor University Medical Center Proc. (2009) 22:287–91. doi: 10.1080/08998280.2009.11928535
68. Jiang YN, Muk T, Stensballe A, Nguyen DN, Sangild PT, Jiand PP. Early protein markers of necrotizing enterocolitis in plasma of preterm pigs exposed to antibiotics. Front Immunol. (2020) 11:565862. doi: 10.3389/fimmu.2020.565862
69. Brunse A, Peng Y, Li Y, Lykkesfeldt J, Sangild PT. Co-bedding of preterm newborn pigs reduces necrotizing enterocolitis incidence independent of vital functions and cortisol levels. Front Pediatr. (2021) 9:636638. doi: 10.3389/fped.2021.636638
70. Birck MM, Nguyen DN, Ciliebord MS, Kamal SS, Nielson DS, Dambord P, et al. Enteral but not parenteral antibiotics enhance gut function and prevent necrotizing enterocolitis in formula-fed newborn preterm pigs. Am J Physiol Gastrointest Liver Physiol. (2016) 310:G323–333. doi: 10.1152/ajpgi.00392.2015
Keywords: intestinal microbiota, translational models, pig models, gut commensals, gastrointestinal disease
Citation: Rose EC, Blikslager AT and Ziegler AL (2022) Porcine Models of the Intestinal Microbiota: The Translational Key to Understanding How Gut Commensals Contribute to Gastrointestinal Disease. Front. Vet. Sci. 9:834598. doi: 10.3389/fvets.2022.834598
Received: 13 December 2021; Accepted: 28 February 2022;
Published: 25 March 2022.
Edited by:
Mark Gray, University of Edinburgh, United KingdomReviewed by:
Bert Devriendt, Ghent University, BelgiumHarry D. Dawson, Agricultural Research Service (USDA), United States
Copyright © 2022 Rose, Blikslager and Ziegler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amanda L. Ziegler, Amanda_ziegler@ncsu.edu
 Elizabeth C. Rose
Elizabeth C. Rose Anthony T. Blikslager
Anthony T. Blikslager Amanda L. Ziegler
Amanda L. Ziegler