Peer-review started: October 27, 2017
First decision: November 23, 2017
Revised: December 6, 2017
Accepted: December 13, 2017
Article in press: December 13, 2017
Published online: February 12, 2018
To identify cell culture models supportive for Zika virus (ZIKV) replication.
Various human and non-human cell lines were infected with a defined amount of ZIKV Polynesia strain. Cells were analyzed 48 h post infection for the amount of intracellular and extracellular viral genomes and infectious viral particles by quantitative real-time PCR and virus titration assay. The extent of replication was monitored by immunofluorescence and western blot analysis by using Env and NS1 specific antibodies. Innate immunity was assayed by luciferase reporter assay and immunofluorescence analysis.
All investigated cell lines except CHO cells supported infection, replication and release of ZIKV. While in infected A549 and Vero cells a pronounced cytopathic effect was observed COS7, 293T and Huh7.5 cells were most resistant. Although the analyzed cell lines released comparable amounts of viral genomes to the supernatant significant differences were found for the number of infectious viral particles. The neuronal cell lines N29.1 and SH-SY5Y released 100 times less infectious viral particles than Vero-, A549- or 293T-cells. However there is no strict correlation between the amount of produced viral particles and the induction of an interferon response in the analyzed cell lines.
The investigated cell lines with their different tissue origins and diverging ZIKV susceptibility display a toolbox for ZIKV research.
Core tip: In this study ten different cell lines, human and non-human, from various tissues (e.g., hepatocytes, keratinocytes and neuronal cells) were tested upon their susceptibility to Zika virus (ZIKV) infection. Except CHO cells all cells supported ZIKV life cycle, but differed in parts strongly in the intracellular and released amount of infectious viral particles. Investigating the interferon response showed no clear correlation between high and low producer cell lines.
- Citation: Himmelsbach K, Hildt E. Identification of various cell culture models for the study of Zika virus. World J Virol 2018; 7(1): 10-20
- URL: https://www.wjgnet.com/2220-3249/full/v7/i1/10.htm
- DOI: https://dx.doi.org/10.5501/wjv.v7.i1.10
The Zika virus (ZIKV) is known since 1947 when it was isolated from a rhesus macaque monkey in a yellow fever research institute in the Zika forest of Uganda[1]. ZIKV has reached global attention during the epidemic in Brazil in the years 2015/2016. This mosquito-borne virus (Aedes aegypti and Aedes albopictus) was found to circulate only in East and West Africa[2] until a bigger outbreak occurred on the Yap Islands in Micronesia in the year 2007[3]. Another outbreak took place in French Polynesia in the year 2013. Here for the first time the congenital ZIKV syndrome (CZVS), microcephaly, the Guillain-Barré syndrome (GBS) and non-vectorborne transmission (mother to child, sexual, posttransfusion) was retrospectively documented[4-7]. However, the virus came into public focus in the beginning of 2016, when the WHO declared the Public Health Emergency of International Concern (PHEIC) since in context of the Brazil epidemic (WHO Zika Strategic Response Plan 2016) a clear correlation between ZIKV infection of pregnant women and fetal microcephaly development was observed[8,9]. This changed the attention from a side note to a headline, initiating a variety of research efforts to investigate the virus in more detail with respect to epidemiology, virus-associated pathogenesis and virus cell interaction.
ZIKV belongs to the Flaviviridae family, which is closely related to the Spondweni virus serocomplex. As member of the Flavivirus genus, ZIKV contains a single-stranded RNA with positive polarity. The viral genome encodes a single polyprotein processed by host and viral proteases into three structural proteins - core (C) that forms the capsid, the precursor of the membrane protein (prM), and the envelope protein (E) - and into seven nonstructural proteins NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 that are responsible for the replication of the viral RNA[10]. The virus replication and morphogenesis occurs in the extranuclear compartment. In ZIKV-infected cells a massive remodeling of the endoplasmic reticulum (ER) to form membranous replication factories and a drastic reorganization of microtubules and intermediate filaments can be observed[11].
There is a variety of reports describing the infection of various primary cells or immortalized cell lines. Moreover, ZIKV was shown to replicate in various human cell types already like skin cells[12] and lung epithelial cells[13]. Not unexpected was the finding that the Aedes C6/36 cells were infectable[12], since this was described for other related viruses already[14]. Furthermore, a lot of animal cell lines were described to be susceptible to ZIKV infection[15]. When mice lacking receptors for IFN-α/β (A129) were infected with the ZIKV, viral RNA could be found in the brain, ovary, spleen and liver[16].
In order to further characterize the virus in human cell lines and to identify cell culture systems that allow the robust production of high amounts of infectious viral particles, ten cell lines were comparatively analyzed for their susceptibility to the ZIKV. Keratinocytes (HaCaT) were included in the following experiments, since the skin is the first tissue the virus comes in contact with via mosquito bite. Moreover neuronal cells (N29.1 and SH-SY5Y) were of special interest due to the neurological disorders ZIKV infections may cause. Furthermore the infectivity of the well-established standard cell lines 293T cells, CHO cells, Vero cells, A549 cells, HepG2C3A cells, Huh7.5 cells and COS7 cells was studied.
A549, CHO, COS7, HepG2/C3A, Huh7.5, HaCaT, N29.1, SH-SY5Y, Vero and 293T cells (Table 1) were grown in Dulbecco’s modified eagle medium (DMEM) supplemented with 2 mmol/L L-Glutamine, non-essential amino acids, 100 U/mL penicillin and 100 μg/mL streptomycin in a humidified incubator at 37 °C with 5% CO2. Passaging of the cells was carried out three times a week, reaching a maximum density of 90%.
| Cell line | Species | Tissue | Origin |
| A549 | Homo sapiens | Lung epithelial | 1972; D. Giard |
| Human | |||
| CHO | Cricetulus griseus | Ovarie | 1957; T. Puck |
| Chinese Hamster | |||
| COS7 | Chlorocebus aethiops | Kidney | 1964; F. Jensen |
| African Green Monkey | |||
| HepG2/C3A | Homo sapiens | Liver | 1980; B. Knowels |
| Human | |||
| Huh7.5 | Homo sapiens | Liver | 1980; B. Knowels |
| Human | |||
| HaCaT | Homo sapiens | Skin | 1988; P. Boulkamp |
| Human | |||
| N29.1 | Mus musculus | Hypothalamus | Cedarlane Laboratories |
| Mouse | |||
| SH-SY5Y | Homo sapiens | Bone marrow Neuroblastoma | 1973; J. Biedler |
| Human | |||
| Vero | Chlorocebus aethiops | Kidney | 1962; Y. Yasumura |
| African Green Monkey | |||
| 293T | Homo sapiens | Kidney | 1973: van der Eb |
| Human |
The cells were infected with the ZIKV strain French Polynesia (PF13/251013-18) (this clinical low passage strain was kindly provided by Professor Musso, Institute Louis Marlade in Papeete, Tahiti).
The inoculum for the infection experiments was derived from Vero cells that were infected for 72 h with ZIKV Polynesia. The obtained cell culture supernatant was filtrated and characterized by titration using plaque assays. Defined aliquots were stored at -80 °C. The cell lines were infected with ZIKV at a MOI = 0.1 for 16 h. The inoculum was removed, cells were washed with prewarmed PBS, cultivated with medium for 32 h and harvested after 48 h if not stated differently.
At the present stage of knowledge no detailed information about the velocity of the infection process in the different cell culture systems is available. To avoid effects that reflect potential differences in the velocity of the infection process cells were infected for 16 h (overnight) to ensure a high infection level. The inoculum was removed, cells were washed with prewarmed PBS, cultivated with medium for 32 h and harvested after 48 h if not stated differently.
Vero cells were seeded at a density of 3 × 105 cells per well in standard six well plates and infected with cleared, serial dilutions of either cell culture supernatant or cleared cellular lysate 6 h later. Another 2 h later, the inoculum was removed and the cells were washed twice with PBS. Then the cellular monolayers were overlaid with DMEM complete containing 0.4% seaplaque agarose. Four days later, the agarose overlay was removed and the wells were washed with PBS. Afterwards, the cells were fixed with 4% formaldehyde for 10 min and stained with 0.1% crystal violet for plaque visualization.
RNA from total lysate was isolated using peqGOLD TriFast (PEQLAB Biotechnologie GmbH, Germany) according to the manufacturer’s protocol. cDNA synthesis was performed after DNA digest with DNaseI (Promega, Mannheim, Germany), using 4 μg total RNA, RevertAid H Minus Reverse Transcriptase and random primer (Thermo Scientific, Dreieich, Germany) as suggested by the manufacturer.
RNA from cell culture supernatant was isolated using QIAamp viral RNA Mini Kit (Qiagen, Hilden Germany) as described by the manufacturer. However, the elution volume was decreased to 40 μL per sample.
Quantitative real-time PCR (qPCR) from total RNA was performed as described[17]. All relative quantifications were normalized to the amount of RPL27 transcripts. The following primers were used: Zika fwd (5’agatcccggctgaaacactg 3’-bp 1924-1943), Zika rev (5’ttgcaaggtccatctgtccc 3’-bp 1996-1977), ribosomal protein L27 - RPL27 fwd (5’ aaagctgtcatcgtgaagaac 3’) and RPL27 rev (5’ gctgctactttgcgggggtag 3’).
qPCR using RNA isolated from cell culture supernatant was analyzed using Zika LightMix Kit (TIB MOLBIOL, Berlin, Germany) in combination with LightCycler® Multiplex RNA Virus Master (Roche, Mannheim Germany) as described by the companies protocols. In brief, 2.7 μL PCR grade water, 0.25 μL Zika Light Mix, 2 μL Roche Master, 0.05 μL RT Enzyme were mixed with 5 μL purified RNA and measured in the LightCycler 480 or Light cycler 1.2 (Roche, Mannheim Germany) with the following program: (1) RT-Step: 55 °C/5 min; (2) Denaturation: 95 °C/5 min; (3) Cycling (45 times): 95 °C/5 s, 60 °C/15 s, 72 °C/15 s; and (4) Cooling: 40 °C/30 s.
The samples were resolved by sodium dodecyl sulfate-polyacrylamid electrophoresis (SDS-PAGE) at 10% and transferred by semi-dry blotting onto a polyvinylidine difluoride membrane (PVDF) (0.45 μm; Carl Roth, Germany). The membrane was blocked with 5% skim milk solution and then incubated with anti NS1 specific antibody at a 1:1000 dilution (Biofront, United States) overnight. Then the membrane was incubated with a mouse specific secondary antibody coupled with horseradish peroxidase at a 1:2000 dilution (HRP) (GE Healthcare, United Kingdom) and signals were detected with X-ray films (GE Healthcare, United Kingdom). Signals were quantified using ImageJ software.
Immunofluorescence staining was analyzed with a confocal laser scanning microscope (CLSM 510 Meta; Carl Zeiss) and ZEN 2009 software. Cells were fixed with absolute ice-cold ethanol for 10 min. ZIKV envelope protein was stained using anti Flavivirus Group antigen Antibody (clone D1-4G2-4-15 from Merck-Millipore, Darmstadt Germany) and a polyclonal rabbit antiserum was used to detect STAT1 (Merck AB16951). As secondary antibodies served Alexa 488 and Alexa 546 (Thermo, Darmstadt Germany). Nuclei were stained with DAPI.
Cytolysis was monitored by LDH release assay (Clontech, Japan) and cell viability was assessed by Presto Blue staining (Thermo, Darmstadt Germany) according to the instructions of the manufacturer. Upon cellular damage lactate dehydrogenase (LDH) is released into the cell culture supernatant. This release is indirectly measured based on a calorimetric assay detecting an enzymatically formed formazan product. Presto Blue is a red compound that is taken up by the cells and due to the reducing interior environment turns into a red color that is detectable at 570 nm.
The used cell lines were transfected with the pISREluc construct (Agilent, United States), using polyethylenimine (PEI) directly after infection. In brief, 3 × 105 cells per six well were infected as described and transfected directly after the addition of virus, using a transfection mix containing 1 μg plasmid DNA, 12 μL PEI (1 mg/mL) in a total volume of 150 μL PBS (1/10 total volume of media). The media was changed the next day and cells were analyzed 48 h post transfection. Here for cells were lysed in a passive lysis buffer (25 mmol/L Tris, 2 mmol/L DTT, 2 mmol/L EGTA, 10% glycerol (v/v), 1% TX-100 (v/v), pH 7.5) for 10 min on ice. Afterwards, lysate was cleared by centrifugation at 4 °C and 5000 × g for 10 min and the luciferase activity of the supernatant was measured in 96 well Orion II plate reader (Berthold, Germany) for 10 s after the addition of luciferase buffer (20 mmol/L Tris-HCl pH 7.8, 5 mmol/L MgCl2, 0.1 mmol/L EDTA, 33.3 mmol/L DTT, 470 μmol/L Luciferin, 530 μmol/L ATP). Relative light units were normalized to the total protein amount by Bradford protein assay.
All statistical analyses were performed with Prism GraphPad 7.0, using multiple t tests for determination of P-values. Error bars are displayed as value ± SEM.
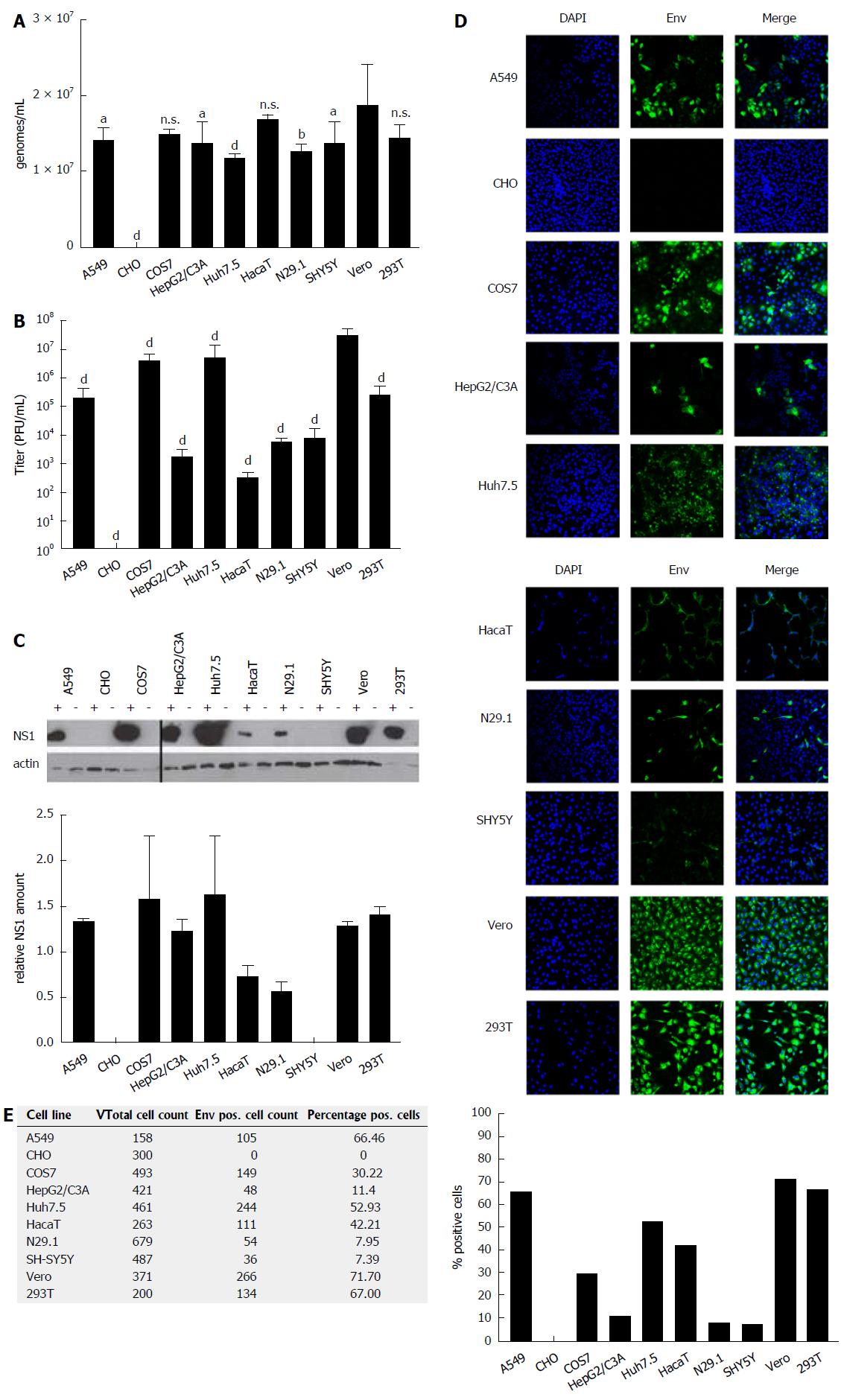
The capacity of various human- and non-human-derived cell lines to produce high amounts of infectious ZIKV particles was analyzed. For this purpose, ten different (human and non-human) cell lines (Table 1) derived from various tissues (neuronal cells, kidney cells, keratinocytes, hepatoma cells and lung epithelia cells) were tested with respect to their susceptibility to ZIKV infection. The investigated cells were infected with an identical MOI of 0.1, using ZIKV Polynesia strain. The intracellular amount of ZIKV-specific genomes was determined by RT-PCR 48 h after infection, revealing that for all analyzed cell lines with the exception of CHO cells a productive infection could be established. With respect to the number of intracellular genomes detected in the various cell lines (Figure 1A), only moderate differences between the permissive cell lines were found. However, the amount of viral genomes must not necessarily correlate with the amount of infectious viral particles. To address this point, the number of infectious particles in the cell lysates was determined. Quantification of the intracellular amount of infectious viral particles by plaque assay (Figure 1B) revealed strong differences between the investigated cell lines of up to 105-fold. The highest amount of intracellular infectious viral particles was found for Vero cells containing 3.6 × 107 PFU/mL followed by the Huh7.5, COS7, 293T and A549 cells. Again, N29.1 and SH-SY5Y cells showed significantly lower amounts of intracellular viral particles (8.7 × 103 - 2.3 × 103 PFU/mL). The HaCaT cells showed besides the CHO cells, which did not contain infectious viral particles, the lowest amount amongst the investigated cell lines (3 × 102 PFU/mL). Comparison between the quantification of the intracellular viral genomes and the infectious viral particles revealed that there is a correlation, but the differences between the various cell lines are much more pronounced with respect to the amount of infectious viral particles in comparison to the viral genomes.
Quantification of intracellular viral genomes does not automatically reflect replication. To further analyze ZIKV replication, the intracellular amount of NS1 was determined by western blot analysis and referred to the amount of actin (Figure 1C). The quantification of the western blots demonstrates that between the different cell lines significant differences with respect to the intracellular amount of NS1 can be observed. Nearly the same pattern for the amount of NS1 can be observed as found for the intracellular genomes by RT-PCR. A549, COS7, HepG2/C3A, Huh7.5, Vero and 293T cells showed strongest signals, while lower amounts of NS1 were detected in N29.1 cells. No NS1 was measurable in SHY5Y and CHO cells. For the HaCaT cells in contrast to the qPCR data only a low amount of NS1 was observed.
To estimate the intracellular amount of ZIKV envelope protein and to analyze the amount of infected vs non-infected cells, confocal immunofluorescence microscopy was performed (Figure 1D). The staining showed that based on the plaque assays the highest producer cells also showed the best ratio between infected vs non-infected cells (Figure 1E). The confocal immunofluorescence microscopy shows for the A549, Vero and 293T cells a susceptibility between 72%-66%, while from the Huh7.5 and HaCaT cells approximately 52% and 42% and only around 10% of the HepG2/C3A, N29.1 and SH-SY5Y were infected after 48 h. In case of the CHO cells, no specific staining was observed confirming that these cells are not permissive for ZIKV.
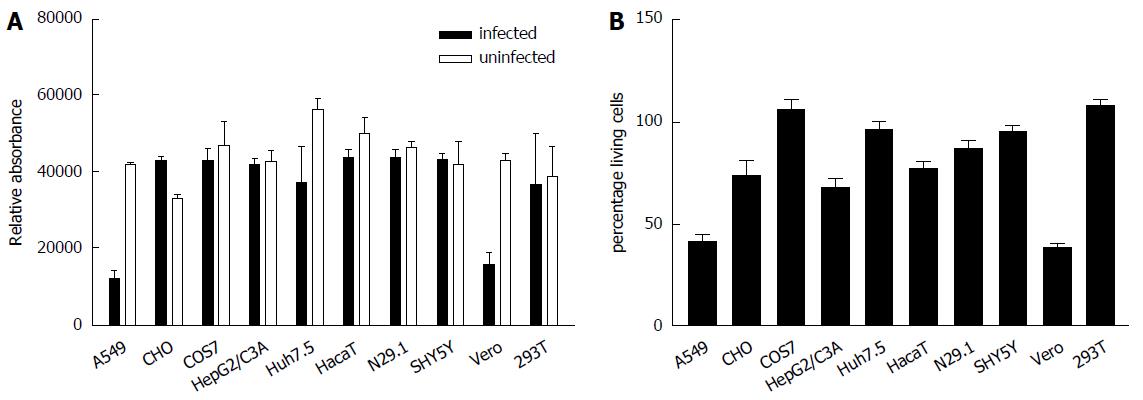
To study the impact of ZIKV on cell viability and integrity in the different analyzed cell lines Presto Blue assays for determination of the cell viability and LDH assays for analysis of the cell integrity were performed. For this purpose the cells were infected for 48 h and stained for Presto Blue assays or the supernatant from ZIKV-infected cells was collected 48 h after infection and the LDH activity was determined (Figure 2). Both assays revealed that ZIKV heavily affects cell integrity/cell viability in A549 and Vero cells. Less cell death was observed for HepG2/C3A, HaCaT and N29.1 cells. Based on the data from these assays and microscopic analysis COS7, 293T and SH-SY5Y cells were found to be most resistant to ZIKV induced cytolysis.
To investigate whether the data obtained for the analysis of the intracellular amount of viral genomes and infectious viral particles are reflected by the numbers of genomes and infectious viral particles in the supernatant, media from the infected cell cultures were analyzed 48 h after infection. The RT-PCR (Figure 3A) revealed that in accordance to the results obtained for the quantification of the intracellular genomes, CHO released no viral genomes. As observed for the intracellular amount of viral genomes, there were no major differences in the amount of released viral genomes between the different cell lines. The difference between the highest amount observed for A549, HaCaT and Vero cells on the one side and N29.1 or SH-SY5Y cells on the other side is less than 2 fold. As the amount of viral genomes in the supernatant must not correlate with the amount of infectious viral particles, the number of infectious viral particles in the cell culture supernatants was determined by virus titration assays (Figure 3B). In contrast to the moderate differences that were found analyzing the number of viral genomes, strong differences (more than 102-fold) were revealed with respect to the number of infectious viral particles released by the different cell lines. The highest amounts were detected for supernatants derived from Vero-, A549-, COS7-, HepG2/C3A-, Huh7.5-, HaCaT- and 293T-cells that produced nearly the same quantity of infectious viral particles (about 107/mL). The neuronal cell lines N29.1 and SH-SY5Y cells released more than 100 times less infectious viral particles than Vero cells. For CHO cells no significant amount of released viral particles was detectable.
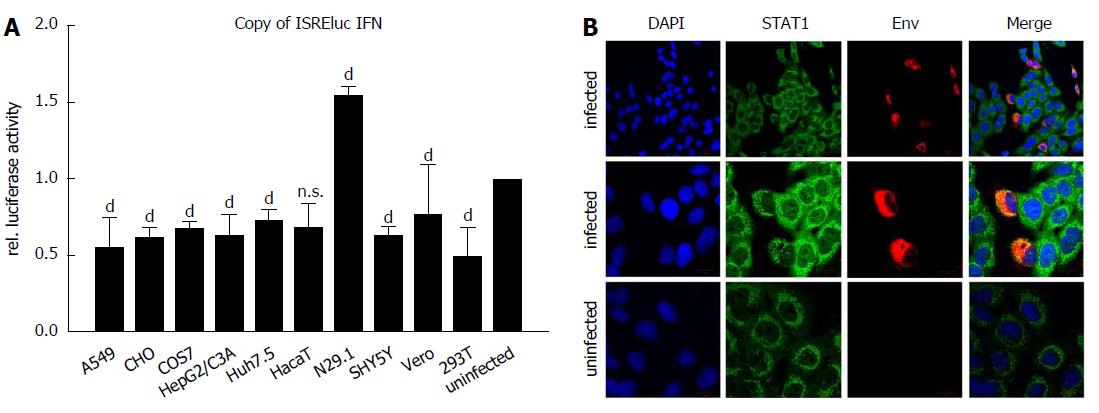
Induction of interferon-stimulated genes (ISGs) was analyzed by luciferase reporter assay using the Interferon-stimulated response element (ISRE) as promoter-driving luciferase expression. Cells were infected as described, transfected with pISREluc plasmid and the cellular luciferase activity was analyzed 48 h post infection (Figure 4A). Luciferase assay showed an induction of ISGs only for the N29.1 cells. In the rest of the tested cells ISGs were slightly repressed. However, staining of STAT1 suggests a delocalization by ZIKV (Figure 4B). If ZIKV was present in the cells it occurred like STAT1 is drawn to the replication factories and no longer is evenly distributed as seen in uninfected cells.
Taken together, these data indicate that the analyzed cell lines strongly differ with respect to the amount of released viral particles, although comparable amounts of viral genomes are detectable in the supernatant. With respect to the identification of cell culture systems that are suitable to produce high amounts of infectious viral particles, Vero- and COS7 cells as a non-human-derived cell lines and Huh7.5-, A549 and 293T cells as human-derived cell culture systems were identified.
ZIKV first isolated from a sentinel monkey in the Zika forest in Uganda in 1947[1] is an emerging virus that has spread over the years from Africa, over Asia, Micronesia, French Polynesia to Brazil[18]. Since the Brazil epidemic in 2015/2016[19] ZIKV research has increased dramatically. In this study a ZIKV isolate from Polynesia that also belongs to the Asian lineage like the ZIKV strain causative for the epidemic in Brazil, was used to characterize its potential to infect various human cell lines with the aim to provide cell culture models for investigating the ZIKV life cycle in more detail and to test the suitability of various cell culture systems to produce high amounts of this virus[20]. The French Polynesia strain was utilized since this was described first to cause GBS and microcephaly, like later for the Brazil outbreak[4,7,20].
In light of the correlation between ZIKV infection of pregnant women and the development of microcephaly in the fetus it is of major interest that ZIKV can be detected in the maternal decidua, fetal placenta and umbilical cells.
The virus targets different cell types such as macrophages, fibroblasts, trophoblasts as well as mesenchymal stem cells[21].
In interferon receptor type I-deficient mice (IFNAR KO) ZIKV causes systemic infections in all tissues providing a useful tool for drug target testing[16,22-24]. Moreover, several publications have shown to replicate ZIKV in various primary cells like neuronal cells, dendritic cells or keratinocytes[12,14,25,26]. Immortalized cells are already described for infection with ZIKV[11,12,14,27].
In this comparative study, ten cells lines were tested for their capacity to support replication and production of infectious virus. After infection of the cells, the ongoing replication was monitored by qPCR, western blot and immunofluorescence microscopy analysis, while the capacity to produce infectious virus was investigated by qPCR and plaque assay. It was found that ZIKV replicates in neuronal cells, keratinocytes, lung carcinoma cells, liver carcinoma cells and kidney cells. Vero cells always served as positive control and standard for quantification. ZIKV showed best replication and virus production in A549, COS7, Huh7.5 and 293T cells, followed by HepG2/C3A and HaCaT cells. Significantly lower support was measured for N29.1 and SH-SY5Y cells. In contrast to these cells, CHO cells could not be infected. The finding that 293T cells are susceptible to ZIKV stands in contrast to a previous publication by Hamel et al[12] (2015). Huh7 cells have also been used in some studies and demonstrated to support viral replication[11], but here we used the Huh7-derived Huh7.5 cells clone[28] that has a defect in the RIG-I gene[29]. However, comparable titers were reached for the Huh7 cells measured by Cortese et al[11] and the Huh7.5 cells in our hands, so that there seems to be no benefit using the Huh7.5 cells at least for the production of high titer viral stock, indicating that ZIKV replication is not significantly affected by the functionality of the RIG-I gene. ZIKV propagation in primary keratinocytes has been shown already[12]. Here we used the immortalized keratinocyte cell line HaCaT[30] which turned out to be susceptible to viral infection and also produced a good viral titer. Moreover, the virus replicates very efficiently in COS7 cells[31], showing release of infectious virus to a titer of over 107 viral particles/mL. This was not unexpected since these cells, like Vero cells, derive from the kidney of an African green monkey. By using a MOI of 0.1, which is in the lower range if compared to Dengue virus titers used for infections in cell culture, we assured to only identify cells that support viral infection efficiently. The measured viral genomes in the corresponding supernatants resembled the findings from the intracellular quantification, in which the measured genomes in case of CHO cells represent input RNA from the infection procedure. However, comparable amounts of viral RNA were observed inside and outside of the cells.
When comparing the intracellular and the extracellular titers of infectious virus, in general higher titers were measurable in the supernatant of the cells compared to the intracellular amount of virus. But for the high producer cell lines like A549, COS7, Huh7.5, Vero and 293T nearly the titers measured outside the cells were reached already inside the cells. This is also reflected by the detection of NS1 in cellular lysate by western blot and by analyzing the Env-protein by immunofluorescence. Here also more NS1 was detectable for the high producers and more infected cells were visualized. However, for unknown reasons only low titers were detected inside the HepG2/C3A cells, but nearly 104 more infectious virus was secreted by these cells. This enhancement of viral release may be based on efficient packaging and export of the viral cargo or a lower turnover of viral proteins.
To investigate if the activation of IFN-signaling is causative for the diverging susceptibility of the analyzed cell lines, the induction of ISGs was monitored by pISRE-luciferase reporter experiments upon viral infection. The luciferase assay showed for all infected cell lines a reduction on ISRE activity, except for N29.1 cells. Inhibition of Type I and II IFN production by interfering with the STAT signaling has been demonstrated in several recent publications for the ZIKV[32-34]. Particularly NS5 is described to counteract IFN signaling by binding STAT2 and to promote STAT2 degradation by the proteasome[32]. Moreover, NS1, NS4B and NS2B3 were also shown to inhibit IFN signaling[34]. In contrast to wt mice having a functional adaptive immune response, IFNAR KO mice are susceptible to ZIKV infection[16,23]. This also emphasizes the importance of the IFN response which has to be trapped by the virus in order to establish infection. Astonishingly, the ISRE promoter element was also reduced in its activation in CHO cells, although they do not support viral infection and replication. Since it is not known if the lack of susceptibility of CHO cells for ZIKV infection is due to impaired attachment, entry or post entry steps there exist a variety of possibilities that could lead to an interference with the interferon signaling. The results from the luciferase assay were strengthened by STAT1 staining in infected A549 cells. The changed distribution in infected vs uninfected cells is obvious, but from these experiments it stays uncertain if this is causative for the reduced ISRE activation. The results of this study support ZIKV research by providing different cell culture models based on various tissues, so that the information at hand enables the investigation of ZIKV life cycle in more detail. Also drug testing and pathogenicity studies can be fostered by the shown cell culture models susceptible to ZIKV.
Zika virus (ZIKV) is an emerging virus transmitted mainly by mosquitos, that has spread during the last decades from Africa, to Asia, over Micronesia to the Americans causing an epidemic in Brazil in the years 2016/2017. In order to propagate the virus in cell culture we investigated various cell lines for their susceptibility to ZIKV infection.
To date ZIKV is mainly propagated in Vero cells derived from kidney epithelial cells from African green monkey. This study aimed to investigate the potential of various cell lines to support the viral life cycle in order to provide researchers with suitable cell culture systems for different issues in the field of ZIKV research.
The objectives of this research were to investigate ten human and non-human cell lines from various tissues (e.g., hepatocytes, keratinocytes and neuronal cells) with regard to their intracellular amount of viral genomes and infectious viral particles upon ZIKV-infection. Moreover, the amount of secreted viral genomes and infectious viral particles was analyzed in the cell culture supernatants. Furthermore, the amount of infected cells was analyzed by immunofluorescence using an Envelope-specific antibody and the amount of NS1 was analyzed by western blot. In order to draw a conclusion whether parts of the innate immune response are responsible for the found differences in viral support, STAT1 distribution and expression was analyzed.
Quantification of viral genomes was performed by qPCR. For the detection of genomes from whole cell lysate a standard PCR protocol with SYBR green was used with cDNA as template transcribed from total RNA that was isolated with a Tri-reagent. Viral genomes released into the cell culture supernatant were isolated with a viral RNA isolation kit and subjected to a Taqman-PCR based on a ZIKV-Lightmix Kit. The detection of infectious virus was performed by virus titration assay using serial dilutions from the supernatant or from cleared cellular lysates. The amount of infected cells was analyzed with immunofluorescence microscopy by using an Env-specific antibody and with western blot using NS1-specific antibody. The effect on the innate immunity was monitored by luciferase-reporter assay and STAT1 distribution was analyzed by immunofluorescence microscopy.
All investigated cell lines except CHO cells supported infection, replication and release of ZIKV. While in infected A549 and Vero cells a pronounced cytopathic effect was observed COS7, 293T and Huh7.5 cells were most resistant. Although the analyzed cell lines released comparable amounts of viral genomes to the supernatant significant differences were found for the number of infectious viral particles. The neuronal cell lines N29.1 and SH-SY5Y released 100 times less infectious viral particles than Vero-, A549- or 293T-cells. However there is no strict correlation between the amount of produced viral particles and the induction of an interferon response in the analyzed cell lines.
The results presented so far provide a toolbox of cell culture systems for ZIKV research in general. However, the analyzed cells differ strongly with respect to the amount of released viral particles, whereas the amount of genomes amongst the cells in the supernatant and inside of infected cells are more or less equal. This is an important finding, since a lot of research and diagnostic is based on qPCR analysis only.
Further research should aim on the differences of released viral genomes vs released infectious virus. Are there differences in the release pathway? Which pathways are used for viral egress? Why are certain cell lines not succeptible?
We thank Dr. Didier Musso (Director of the Research and Diagnosis Laboratory, Institute Louis Marlade in Papeete, Tahiti) for disposing ZIKV Polynesia strain (PF 13/251013-18).
Manuscript source: Unsolicited manuscript
Specialty type: Virology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Carneiro PM, Cunha C, Diefenbach R, Giannecchini S S- Editor: Ji FF L- Editor: A E- Editor: Li RF
| 1. | Dick GW. Epidemiological notes on some viruses isolated in Uganda; Yellow fever, Rift Valley fever, Bwamba fever, West Nile, Mengo, Semliki forest, Bunyamwera, Ntaya, Uganda S and Zika viruses. Trans R Soc Trop Med Hyg. 1953;47:13-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 89] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Olson JG, Ksiazek TG, Suhandiman , Triwibowo . Zika virus, a cause of fever in Central Java, Indonesia. Trans R Soc Trop Med Hyg. 1981;75:389-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 265] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 3. | Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536-2543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2211] [Cited by in F6Publishing: 2041] [Article Influence: 136.1] [Reference Citation Analysis (0)] |
| 4. | Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, Baudouin L, Mallet H, Musso D, Ghawche F. Zika virus infection complicated by Guillain-Barre syndrome--case report, French Polynesia, December 2013. Euro Surveill. 2014;19:pii: 20720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 676] [Cited by in F6Publishing: 673] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 5. | Calvet G, Aguiar RS, Melo ASO, Sampaio SA, de Filippis I, Fabri A, Araujo ESM, de Sequeira PC, de Mendonça MCL, de Oliveira L. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect Dis. 2016;16:653-660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 767] [Cited by in F6Publishing: 779] [Article Influence: 97.4] [Reference Citation Analysis (0)] |
| 6. | Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jääskeläinen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016;374:2142-2151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 634] [Cited by in F6Publishing: 600] [Article Influence: 75.0] [Reference Citation Analysis (0)] |
| 7. | Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374:951-958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2157] [Cited by in F6Publishing: 1830] [Article Influence: 228.8] [Reference Citation Analysis (0)] |
| 8. | Fauci AS, Morens DM. Zika Virus in the Americas--Yet Another Arbovirus Threat. N Engl J Med. 2016;374:601-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 727] [Cited by in F6Publishing: 650] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 9. | Zhang Q, Sun K, Chinazzi M, Pastore Y Piontti A, Dean NE, Rojas DP, Merler S, Mistry D, Poletti P, Rossi L. Spread of Zika virus in the Americas. Proc Natl Acad Sci USA. 2017;114:E4334-E4343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 171] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 10. | Kuno G, Chang GJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007;152:687-696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 362] [Cited by in F6Publishing: 374] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 11. | Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017;18:2113-2123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 221] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 12. | Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89:8880-8896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 902] [Cited by in F6Publishing: 828] [Article Influence: 92.0] [Reference Citation Analysis (0)] |
| 13. | Frumence E, Roche M, Krejbich-Trotot P, El-Kalamouni C, Nativel B, Rondeau P, Missé D, Gadea G, Viranaicken W, Desprès P. The South Pacific epidemic strain of Zika virus replicates efficiently in human epithelial A549 cells leading to IFN-β production and apoptosis induction. Virology. 2016;493:217-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 14. | White LA. Susceptibility of Aedes albopictus C6/36 cells to viral infection. J Clin Microbiol. 1987;25:1221-1224. [PubMed] [Cited in This Article: ] |
| 15. | Barr KL, Anderson BD, Prakoso D, Long MT. Working with Zika and Usutu Viruses In Vitro. PLoS Negl Trop Dis. 2016;10:e0004931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R. A Susceptible Mouse Model for Zika Virus Infection. PLoS Negl Trop Dis. 2016;10:e0004658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 234] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 17. | Himmelsbach K, Hildt E. The kinase inhibitor Sorafenib impairs the antiviral effect of interferon α on hepatitis C virus replication. Eur J Cell Biol. 2013;92:12-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Hayes EB. Zika virus outside Africa. Emerg Infect Dis. 2009;15:1347-1350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 591] [Cited by in F6Publishing: 530] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 19. | Heukelbach J, Alencar CH, Kelvin AA, de Oliveira WK, Pamplona de Góes Cavalcanti L. Zika virus outbreak in Brazil. J Infect Dev Ctries. 2016;10:116-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 20. | Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. An update on Zika virus infection. Lancet. 2017;390:2099-2109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 415] [Cited by in F6Publishing: 391] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 21. | El Costa H, Gouilly J, Mansuy JM, Chen Q, Levy C, Cartron G, Veas F, Al-Daccak R, Izopet J, Jabrane-Ferrat N. ZIKA virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci Rep. 2016;6:35296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 132] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 22. | Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe. 2016;19:720-730. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 743] [Cited by in F6Publishing: 707] [Article Influence: 88.4] [Reference Citation Analysis (0)] |
| 23. | Ma W, Li S, Ma S, Jia L, Zhang F, Zhang Y, Zhang J, Wong G, Zhang S, Lu X. Zika Virus Causes Testis Damage and Leads to Male Infertility in Mice. Cell. 2016;167:1511-1524.e10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 262] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 24. | Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell. 2016;165:1081-1091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 662] [Cited by in F6Publishing: 639] [Article Influence: 79.9] [Reference Citation Analysis (0)] |
| 25. | Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci USA. 2016;113:14408-14413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 333] [Cited by in F6Publishing: 337] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 26. | Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016;18:587-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 904] [Cited by in F6Publishing: 917] [Article Influence: 114.6] [Reference Citation Analysis (0)] |
| 27. | Chan JF, Yip CC, Tsang JO, Tee KM, Cai JP, Chik KK, Zhu Z, Chan CC, Choi GK, Sridhar S. Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg Microbes Infect. 2016;5:e93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 28. | Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001-13014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 980] [Cited by in F6Publishing: 995] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 29. | Sumpter R Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689-2699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 736] [Cited by in F6Publishing: 701] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 30. | Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761-771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3142] [Cited by in F6Publishing: 3231] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 31. | Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175-182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1652] [Cited by in F6Publishing: 2120] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 32. | Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016;17:1766-1775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 33. | Bowen JR, Quicke KM, Maddur MS, O‘Neal JT, McDonald CE, Fedorova NB, Puri V, Shabman RS, Pulendran B, Suthar MS. Zika Virus Antagonizes Type I Interferon Responses during Infection of Human Dendritic Cells. PLoS Pathog. 2017;13:e1006164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 196] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 34. | Wu Y, Liu Q, Zhou J, Xie W, Chen C, Wang Z, Yang H, Cui J. Zika virus evades interferon-mediated antiviral response through the co-operation of multiple nonstructural proteins in vitro. Cell Discov. 2017;3:17006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |