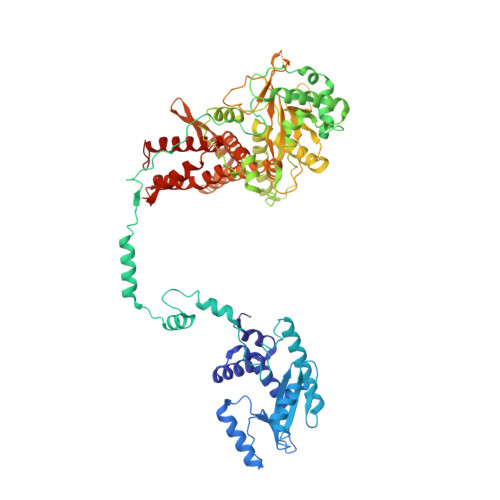
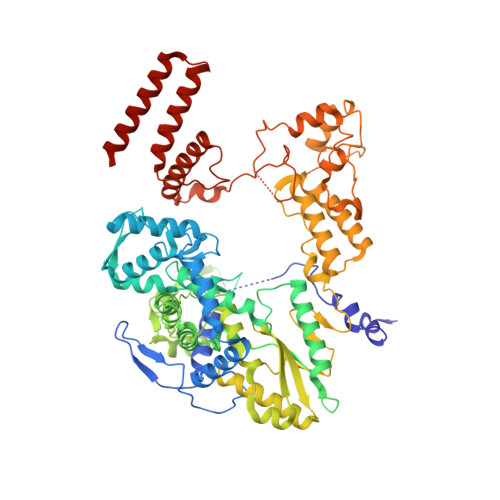
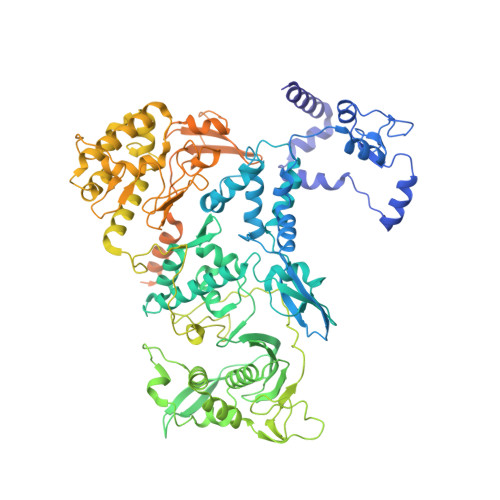
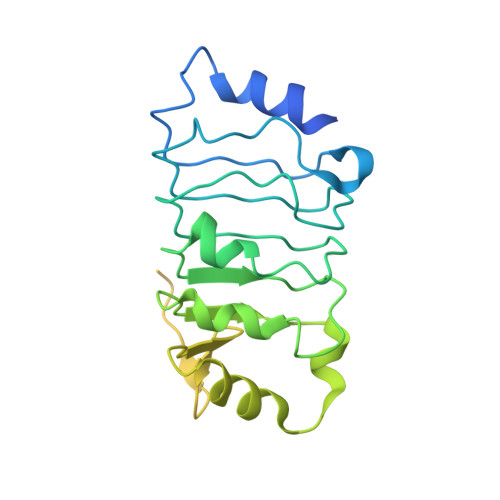
Host ANP32A mediates the assembly of the influenza virus replicase.
Carrique, L., Fan, H., Walker, A.P., Keown, J.R., Sharps, J., Staller, E., Barclay, W.S., Fodor, E., Grimes, J.M.(2020) Nature 587: 638-643
- PubMed: 33208942
- DOI: https://doi.org/10.1038/s41586-020-2927-z
- Primary Citation of Related Structures:
6XZD, 6XZG, 6XZP, 6XZQ, 6XZR, 6Y0C - PubMed Abstract:
Aquatic birds represent a vast reservoir from which new pandemic influenza A viruses can emerge 1 . Influenza viruses contain a negative-sense segmented RNA genome that is transcribed and replicated by the viral heterotrimeric RNA polymerase (FluPol) in the context of viral ribonucleoprotein complexes 2,3 . RNA polymerases of avian influenza A viruses (FluPolA) replicate viral RNA inefficiently in human cells because of species-specific differences in acidic nuclear phosphoprotein 32 (ANP32), a family of essential host proteins for FluPol activity 4 . Host-adaptive mutations, particularly a glutamic-acid-to-lysine mutation at amino acid residue 627 (E627K) in the 627 domain of the PB2 subunit, enable avian FluPolA to overcome this restriction and efficiently replicate viral RNA in the presence of human ANP32 proteins. However, the molecular mechanisms of genome replication and the interplay with ANP32 proteins remain largely unknown. Here we report cryo-electron microscopy structures of influenza C virus polymerase (FluPolC) in complex with human and chicken ANP32A. In both structures, two FluPolC molecules form an asymmetric dimer bridged by the N-terminal leucine-rich repeat domain of ANP32A. The C-terminal low-complexity acidic region of ANP32A inserts between the two juxtaposed PB2 627 domains of the asymmetric FluPolA dimer, suggesting a mechanism for how the adaptive PB2(E627K) mutation enables the replication of viral RNA in mammalian hosts. We propose that this complex represents a replication platform for the viral RNA genome, in which one of the FluPol molecules acts as a replicase while the other initiates the assembly of the nascent replication product into a viral ribonucleoprotein complex.
Organizational Affiliation:
Division of Structural Biology, University of Oxford, Oxford, UK.