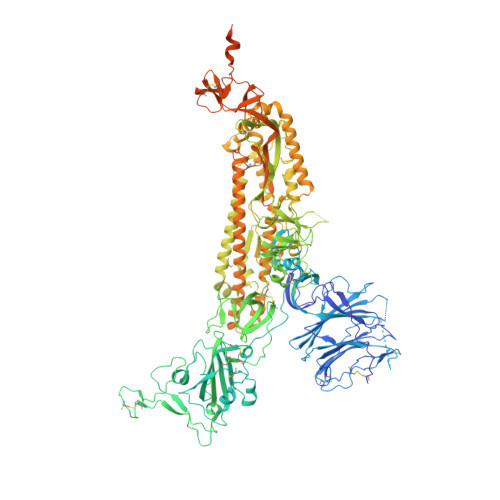
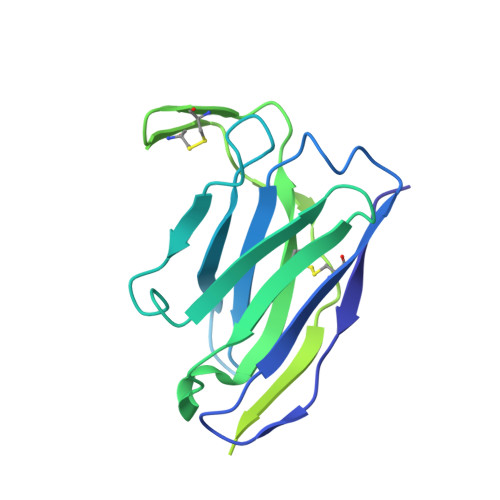
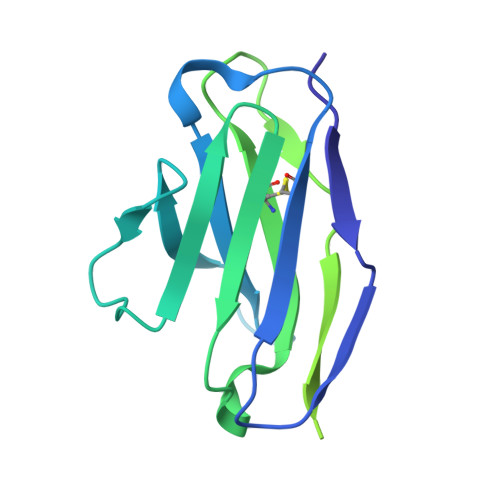
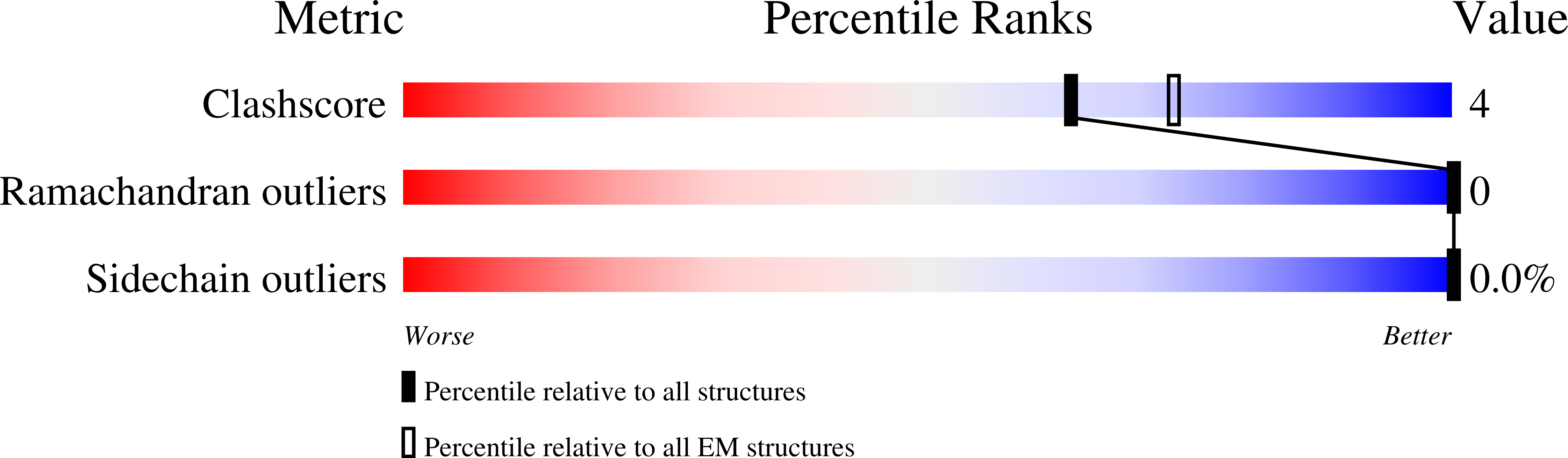Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies.
Cerutti, G., Rapp, M., Guo, Y., Bahna, F., Bimela, J., Reddem, E.R., Yu, J., Wang, P., Liu, L., Huang, Y., Ho, D.D., Kwong, P.D., Sheng, Z., Shapiro, L.(2021) Structure 29: 655-663.e4
- PubMed: 34111408
- DOI: https://doi.org/10.1016/j.str.2021.05.014
- Primary Citation of Related Structures:
7LS9, 7LSS - PubMed Abstract:
Emerging SARS-CoV-2 strains, B.1.1.7 and B.1.351, from the UK and South Africa, respectively, show decreased neutralization by monoclonal antibodies and convalescent or vaccinee sera raised against the original wild-type virus, and are thus of clinical concern. However, the neutralization potency of two antibodies, 1-57 and 2-7, which target the receptor-binding domain (RBD) of the spike, was unaffected by these emerging strains. Here, we report cryo-EM structures of 1-57 and 2-7 in complex with spike, revealing each of these antibodies to utilize a distinct mechanism to bypass or accommodate RBD mutations. Notably, each antibody represented an immune response with recognition distinct from those of frequent antibody classes. Moreover, many epitope residues recognized by 1-57 and 2-7 were outside hotspots of evolutionary pressure for ACE2 binding and neutralizing antibody escape. We suggest the therapeutic use of antibodies, such as 1-57 and 2-7, which target less prevalent epitopes, could ameliorate issues of monoclonal antibody escape.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA; Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY 10029, USA.