Abstract
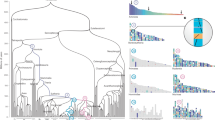
Comparative mapping, which compares the location of homologous genes in different species, is a powerful tool for studying genome evolution1. Comparative maps suggest that rates of chromosomal change in mammals can vary from one to ten rearrangements per million years1,2,3,4. On the basis of these rates we would expect 84 to 600 conserved segments in a chicken comparison with human or mouse. Here we build comparative maps between these species and estimate that numbers of conserved segments are in the lower part of this range. We conclude that the organization of the human genome is closer to that of the chicken than the mouse and by adding comparative mapping results from a range of vertebrates, we identify three possible phases of chromosome evolution. The relative stability of genomes such as those of the chicken and human will enable the reconstruction of maps of ancestral vertebrates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Andersson,L. et al. The first international workshop on comparative genome organisation. Mamm. Genome 7, 717–734 (1997).
Davisson,M. T., Bradt,D. W., Merriam,J. J., Rockwood,S. F. & Eppig,J. T. The mouse gene map. Inst. Lab. Anim. Res. J. 39, 96–131 (1998).
Nadeau,J. H. & Taylor,B. A. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. USA 81, 814–818 (1984).
O'Brien,S. J., Wienberg,J. & Lyons,L. A. Comparative genomics: lessons from cats. Trends Genet. 13, 393–399 (1997).
Waddington,D., Springbett,A. J. & Burt,D. W. A chromosome based model to estimate the number of conserved segments between pairs of species from comparative genetic maps. Genetics (in the press).
Morizot,D. C. Reconstructing the gene map of the vertebrate ancestor. Anim. Biotech. 5, 113–122 (1994).
Sarich,V. M. & Wilson,A. C. Immunological time scale for hominid evolution. Science 158, 1200–1203 (1994).
Novacek,M. J. Mammalian phylogeny: shaking the tree. Nature 356, 121–125 (1992).
de Jong,W. W. Molecules remodel the mammalian tree. Trends Ecol. Evol. 13, 270–275 (1998).
Kumar,S. & Hedges,B. A molecular time-scale for vertebrate evolution. Nature 392, 917–920 (1998).
Watanabe,T. K. et al. A radiation hybrid map of the rat genome containing 5,255 markers. Nature Genet. 22, 27–36 (1999).
Bush,G. L., Case,S. M., Wilson,A. C. & Patton,J. L. Rapid speciation and chromosomal evolution in mammals. Proc. Natl. Acad. Sci. USA 74, 3942–3946 (1977).
O'Neill,R. J. W., O'Neill,M. J. & Graves,J. A. M. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393, 68–72 (1998).
Li,W.-H., Ellsworth,D. L., Krushkal,J., Chang,B. H.-J. & Hewett-Emmett,D. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol. Phylogenet. Evol. 5, 182–187 (1996).
Ohta,T. An examination of the generation-time effect on molecular evolution. Proc. Natl. Acad. Sci. USA 90, 10676–10680 (1993).
Vogel,F., Kopun,M. & Rathenberg, R. in Molecular Anthropology (eds Goodman, M. & Tashian, R. E.) 13–33 (Plenum, New York, 1976).
Burt,D. W. & Cheng,H. H. The chicken gene map. Inst. Lab. Anim. Res. J. 39, 229–236 (1998).
Hastie,T. J. & Tibshirani,R. J. Generalised Additive Models (Chapman and Hall, London, 1990).
Hurvich,C. M., Simonoff,J. S. & Tsai,C.-L. Smoothing parameter selection in non-parametric regression using an improved Akaike information criterion. J. R. Statist. Soc. 60, 271–293 (1998).
Kosambi,D. D. The estimation of map distance from recombination values. Ann. Eugen. 12, 172–175 (1944).
Morton,N. E. Parameters of the human genome. Proc. Natl. Acad. Sci. USA 88, 7474–7476 (1991).
Wienberg,J. & Stanyon,R. Comparative chromosome painting of primate genomes. Inst. Lab. Anim. Res. J. 39, 77–91 (1998).
Acknowledgements
We thank members of the EC ChickMAP project and H. Cheng for providing data prior to publication, and J. Burt for helpful comments. Genome research at the Roslin Institute is supported by the Ministry of Agriculture, Fisheries and Food, the Biotechnology and Biological Sciences Research Council and the Commission of the European Communities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Burt, D., Bruley, C., Dunn, I. et al. The dynamics of chromosome evolution in birds and mammals. Nature 402, 411–413 (1999). https://doi.org/10.1038/46555
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/46555
This article is cited by
Ultra-fast genome comparison for large-scale genomic experiments
Scientific Reports (2019)
Central regulation of feeding behavior through neuropeptides and amino acids in neonatal chicks
Amino Acids (2019)
An adaptable chromosome preparation methodology for use in invertebrate research organisms
BMC Biology (2018)
Assembly of Schizosaccharomyces cryophilus chromosomes and their comparative genomic analyses revealed principles of genome evolution of the haploid fission yeasts
Scientific Reports (2018)
Integration of molecular cytogenetics, dated molecular phylogeny, and model-based predictions to understand the extreme chromosome reorganization in the Neotropical genus Tonatia (Chiroptera: Phyllostomidae)
BMC Evolutionary Biology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.