- College of Agriculture, Guangxi University, Nanning, Guangxi, China
The fruit peel color is an important factor that affects its quality. However, genes involved in regulating pericarp color in bottle gourd (Lagenaria siceraria) have not been explored to date. Genetic analysis of color traits in bottle gourd peel through a genetic population of six generations demonstrated that the green color of peels is inherited as a single gene dominant trait. Combined phenotype-genotype analysis of recombinant plants using BSA-seq mapped the candidate gene to a 22.645 Kb interval at the head end of chromosome 1. We observed that the final interval contained only one gene, LsAPRR2 (HG_GLEAN_10010973). Sequence and spatiotemporal expression analyses of LsAPRR2 unraveled two nonsynonymous mutations (A→G) and (G→C) in the parental CDS sequences. Further, LsAPRR2 expression was higher in all green-skinned bottle gourds (H16) at various stages of fruit development than in white-skinned bottle gourds (H06). Cloning and sequence comparison of the two parental LsAPRR2 promoter regions indicated 11 bases insertion and 8 SNPs mutations in the region -991~-1033, upstream of the start codon in white bottle gourd. Proof of GUS reporting system, Genetic variation in this fragment significantly reduced the expression of LsAPRR2 in the pericarp of white bottle gourd. In addition, we developed a tightly linked (accuracy 93.88%) InDel marker for the promoter variant segment. Overall, the current study provides a theoretical basis for comprehensive elucidation of the regulatory mechanisms underlying the determination of bottle gourd pericarp color. This would further help in the directed molecular design breeding of bottle gourd pericarp.
Introduction
The bottle gourd [Lagenaria siceraria (Mol.) Standl.] (2n = 2x = 22) is native to India and Africa (Whitaker et al., 1971; Meggers et al., 1973; Erickson et al., 2005), and it is cultivated throughout China. It is also widely cultivated in tropical to temperate regions worldwide. The pericarp color is one of the most important qualities influencing consumers’ preferences. Diverse phenotypic variations in bottle gourd peel color include dark green, green, light green, white, etc. The color of the peel mainly depends on the content and form of pigments (Hao et al., 2018). However, the genetic determinants of bottle gourd peel color trait and the underlying regulatory mechanisms have not yet been studied to date.
Previously, several studies have attempted to unravel the molecular patterns of cucurbits peel color inheritance to accelerate the selective breeding process. Several studies have uncovered the genes involved in the regulation of pericarp color trait in some Cucurbitales. In melons, the gene MELO3C003375, which controls fruit skin color, was identified and compared among fruits with different peel color. It was observed that the CDS sequences has a 13bp insertion mutation in the fruits with yellow and white epidermal cells, which is responsible for variation in the peel color of melon (Ou, 2019). Feder et al. (2015) identified an F-box protein-encoding gene (CmKFB) containing a Kelch structural domain in melons. Zhao et al. (2019) Genome-wide association analysis using 635 germplasm demonstrated that the MELO3C003375 (APRR2) on chromosome 4, and the CmKFB gene on chromosome 10 are the major regulators of green and yellow peel colors in melon. Dong et al. (2012) found that the white pericarp trait in cucumber is controlled by the cryptic single gene w. Further, Lun et al. (2016) reported that a Ycf54-like protein encoded by the Csa6G133820 gene is associated with the cyclase activity in the chlorophyll biosynthesis pathway in cucumber. In addition, Liu et al. (2016) identified a candidate gene, APRR2, affecting the green color of the pericarp in cucumber. In white pericarp, the single base insertion of w gene led to the premature appearance of stop codon, which disabled the alleles involved in chlorophyll accumulation and chloroplast development. Transcriptome analysis of the transgenic lines overexpressing the APPR2-Like gene in tomato and a homolog of APPR2-Like gene in sweet pepper (Capsicum annuum) revealed that both genes were associated with pigment accumulation in fruit (Pan et al., 2013). A novel gene, SG1, was found to be highly expressed in young green tissues during a mutagenesis study in Arabidopsis. In the mutants, protoplast to chloroplast transition was delayed, thereby affecting the expression of several genes related to chloroplast development, photosynthesis, and chlorophyll biosynthesis. Overall, the study demonstrated that the protein SG1 is essential for chloroplast development in Arabidopsis (Hu et al., 2014). Identification of Csa7G051430, a candidate gene encoding the (ARC5) protein, plays a critical role in the chloroplast division in Arabidopsis (Zhou et al., 2015). In wax gourd, Ma et al. (2021) reported a candidate gene Bch05G003950 (BhAPRR2) that controls peel color. In white peel wax gourd, a two-base (GA) deletion in the coding sequence of BhAPRR2 with a premature stop codon affects the color of the rind. In a previous study focusing on the color of light and dark green striped watermelon, Elad et al. (2019) observed a mutation at the junction of intron 6 and exon 7, (AG→AC) in the CLAPRR2 gene that controls peel color. The mutation resulted in a 16bp deletion in the coding region of the CLAPRR2 gene in light green striped watermelon, triggering premature transcriptional termination, which might be linked to the light-colored rind in watermelon. In addition, Li et al. (2019) also identified a candidate gene ClCG08G017810 (ClCGMenG) associated with watermelon peel color.
Variations in the chlorophyll, carotenoid, and flavonoid content are responsible for color differences between varieties (Li et al., 2007; Zhu et al., 2016). Several key transcription factors affecting the fruit’s peel color have been identified previously. MYBs are a large family of transcription factors that mainly regulate the secondary metabolite pathways such as phenylpropanoid metabolism, flavonoid biosynthesis, and anthocyanin biosynthesis in plants (Ma et al., 2021). Studies have shown that MYB-like transcription factors are involved in the regulation of the formation of plant anthocyanins, which play an important role in the coloration of fruit skin, flesh, and leaves (Takos et al., 2006; Azuma et al., 2008; Qing et al., 2009). The GOLDEN2-like (GLK) gene, a member of the GARP subfamily of MYB transcription factors, regulates chloroplast development in different plant species (Fitter et al., 2002). For example, regulation of chlorophyll expression levels by the GOLDEN2-like (GLK) transcription factor can affect the fruit color in pepper (Capsicum annuum L.) (Brand et al., 2014), tomato (Solanum lycopersicum L) (Powell et al., 2012; Nguyen et al., 2014), and Arabidopsis (Arabidopsis thaliana) (Fitter et al., 2002; Ni et al., 2017). Some plants require the GOLDEN2-LIKE (GLK) transcription factor for chloroplast development. Waters et al. (2009) showed that GLK proteins regulate the genes involved in light uptake and chlorophyll biosynthesis through direct interaction with promoter sequences in Arabidopsis, and it may optimize photosynthesis by coordinating the plant responses to the environment. In kiwifruit (Actinidia chinensis) (Li et al., 2018), and maize (Zea mays L) (Rossini et al., 2001), GLK2 was found to affect chloroplast compartment size during immature fruit development. GLK2 is shown to be a conserved and important TF that regulates chloroplast development in fleshy fruits in a variety of plants (Jia et al., 2020).
The KNOX family plays a key role in chlorophyll expression in plant fruits. Nadakuduti et al. (2014) showed that TKN2 and TKN4 belong to the class I KNOX family. TKN2 and TKN4 act upstream of SlGLK2 and its related gene SlAPRR2-LIKE, thus affecting the development of chloroplasts in fruit. TKN4 is responsible for regulating the expression of SlGLK2. Altogether, TKN2 and TKN4 regulate the expression of GLK2 and APRR2 (Nadakuduti et al., 2014; Jia et al., 2020).
APRR2 is a transcription factor widely involved in chlorophyll biosynthesis in fruit. Pan et al. (2013) identified a transcription factor in tomato sharing homology with the APRR2-LIKE gene APRR2-LIKE. Overexpression of this gene in transgenic lines led to increased plastid number, area, and pigment content. While the expression levels of APRR2-Like were significantly higher in the leaves of transgenic tomato than in wild-type, total chlorophyll content was similar in both. Unlike SlGLK2, SlAPRR2-LIKE is thought to play a role in the maturation process (Pan et al., 2013). A putative homolog of the tomato APRR2-like gene in sweet pepper (Capsicum annuum) was associated with pigment accumulation in fruit (Pan et al., 2013). Through map-based cloning, it was found that a single base insertion in the APRR2-Like gene caused the termination codon to appear in advance, which was the reason for the white appearance of cucumber immature fruit (Liu et al., 2016). Recently, APRR2 has been reported to be a candidate gene in melon and watermelon for auxiliary regulation of pigment accumulation (Elad et al., 2019). Thus, APRR2 and GLK2 are mutually independent TFs known to control plants’ chloroplast development (Nadakuduti et al., 2014). These studies have shown that the APRR2-LIKE gene plays an important role in promoting pigment accumulation and chloroplast development in plant fruits (Pan et al., 2013). Given the importance of APRR2 in chloroplast development and pigment accumulation, it is imperative to study the potential molecular mechanism underlying the mode of action of APRR2.
Though several studies focusing on the peel color in the Cucurbitaceae family, the genes involved in regulating peel color in bottle gourd have not been identified to date. In the current study, we used a pair of H16 (green) and H06 (white) parent lines, and F1 and F2 generations were selected for the genetic mining of bottle gourd peel color traits. The BSA-seq combined with the phenotype-genotype analysis of recombinant plants and the sequence alignment followed by qRT-PCR were used to identify the candidate genes affecting bottle gourd peel color. Our study revealed the genetic pattern associated with green and white rind traits in bottle gourd for the first time, which is of great value and significance for studying its potential molecular regulatory mechanism. The outcomes of the current study would further help in developing genetic breeding programs for peel color traits in bottle gourd and other Cucurbitaceae family members.
Materials and methods
Plant material and phenotype statistics
Two bottle gourd high-generation inbred lines were selected as parents, with H06 as the white rind parent and H16 as the green rind parent; a six-generation genetic population was constructed for the genetic study of rind colour. The F1 generation was obtained by crossing H06 with H16 and the F1 generation was self-pollinated to obtain the F2 generation. The F1 was backcrossed with H06 and H16 to obtain BC1P1 and BC1P2 respectively. Parents and F1 planted in spring and autumn 2021 and spring 2022, BC1P1 and BC1P2 planted in spring 2022, 634 F2 preliminary locality population planted in autumn 2021, 5208 F2 finely positioned populations planted in spring 2022. The spacing within the planting rows is 0.75 m and the rows are 1.00 m. During planting, only 1 bottle gourd per plant is kept in order to ensure the healthy development of each fruit to be tested. Bottle gourds for phenotypic measurements are harvested 30 days after pollination (DAP) to ensure stable rind colour and chlorophyll content. The colour of the peel was observed with the naked eye immediately after harvest and recorded as green if it matched the colour of the green-skinned parent and as white if it matched the colour of the white-skinned parent.
Cytological observations and pigment content assays
Microscopic observation: To observe the chloroplast morphology of the cell epidermis, well-developed fruits from both parents of 30 DAP were harvested, the thin layer of parental epidermis was carefully peeled off with a sterile scalpel, the section was placed on a slide, a drop of distilled water was added, the slide was carefully covered along one end, the light microscope was completed and then observed under a 400 × fluorescent microscope BX53 (Olympus, Japan) and photographed.
Transmission electron microscopy(TEM):To observe the ultrastructure of chloroplasts in the pericarp of two parents of 30 DAP. The fruit epidermis was quickly cut into 1-2 mm³ size with a scalpel and immediately fixed in pre-cooled electron microscope fixative (2.5% glutaraldehyde solution), the samples were rinsed with 0.1 M, pH 7.0 phosphate buffer and fixed in 1% OsO4 solution; the samples were rinsed three more times; dehydrated at room temperature and the samples were permeabilised and embedded. The samples were cut into 70-90 nm sections using a LEICA EM UC7 (LEICA, Germany). The sections were stained, dried and the chloroplast structure was observed using a transmission electron microscope Hitachi H-7650 (HITACHI, Japan).
The pericarp pigment content was measured, as described by Ma et al. (2021). After fruit harvesting, a portion of the peel with consistent color was selected, and the peel was quickly scraped, placed into liquid nitrogen, and ground to a fine powder. 0.2 g of the ground tissue was weighed into a 15 ml centrifuge tube and sealed after quickly adding 15 ml absolute ethanol. The extraction was completed by shaking at 200 rpm for 24h on a rotating instrument in a dark environment, followed by centrifugation at 6000 rpm. A microplate reader infinite M200 (TECAN, Switzerland) was used to measure the absorbance values of chlorophyll a, chlorophyll b and carotenoids at 665 nm, 649 nm and 470 nm, respectively. Each experiment was repeated three times. All experiments were performed quickly in low light conditions to reduce pigment degradation.
DNA extraction
As described by Wilkie et al. (1997), CTAB method was used to extract genomic DNA from young leaves of parents and F2 generation. Detection of DNA concentration and purity using an ultramicro spectrophotometer k5800 (Kaiao, Beijing, China), and DNA quality was assessed by electrophoresis on 1.2% agarose gels.
BSA-seq mapping approach
Bulked Segregant Analysis (BSA-seq) is a highly efficient and molecular marker-independent method for rapidly and accurately targeting genes to a compartment. It has become an effective tool for gene-targeting studies (Xu et al., 2019). We selected one green parent (H16) and one white parent (H06) for the study. Thirty extremely green individuals and 30 extremely white individuals were selected from the 634 F2 population for extreme admixture pool construction. Genomic DNA from young leaves was extracted, DNA samples tested for database construction, and re-sequenced using the Illumina HiSeq™ PE150 (San Diego, CA, United States) The original readings obtained from high-throughput sequencing were analyzed and converted into sequencing readings after base calls, and filtered to obtain clean readings for subsequent analysis. After filtering, two pools of extreme trait data were admixed, and two pools of parental data were constructed for association analysis. The resequenced data were compared to obtain variant information using the ZAAS_Lsic_2.0 (Xu et al., 2021) as the reference sequence. Two association analysis methods were used: the Euclidean distance (ED) association algorithm and the ΔSNP/index algorithm. Regions associated with the target traits were determined.
Fine mapping and marker development
The kompetitive allele-specific PCR (KASP) genotyping assay exploits a competitive allele-specific PCR and fluorescence-based reporter system to efficiently identify and measure genetic variation occurring at the nucleotide level (He et al., 2014). Based on the BSA-seq results, we initially designed KASP and InDel Markers to genotype the F2 plants for identifying the recombinant plants (Supplementary Table S2). Analysis according to manufacturer ‘s instructions (LGC Genomics, Shanghai, China). In short, the kompetitive allele-specific PCR reaction contained 1.0 μL DNA (8-15 ng/μL), 1.5 μL 2 × Master Mix, 0.5 μL (10 μm) Primer mixture. Landing PCR was used for amplification. The reaction conditions were: 95°C 15min, 95°C 20s denaturation, 65 ~ 55°C annealing amplification for 1min, (10 cycles, each cycle reduced by 1.0°C), 95°C 20s denaturation, 57°C annealing amplification for 1min, (25 cycles, standard procedure), and then stored in dark conditions at 4°C. PCR products were scanned and genotyped.
The InDel-labeled PCR reaction system contained 2.0 μL genomic DNA (20-50 ng/μL), 1.0 μL each of forward and reverse primers (10 μm), 6.0 μL 2 × PCR Master Mix and 2.0 μL ddH2O. Reaction conditions: 95C° pre-deformation 3min, 95C° denaturation 30s, 55-60C° annealing 30s, 72C° extension 30s, (30 ~ 35 cycles, standard procedure); 72C° extension 5min followed by 4C° dark preservation. PCR amplification products were separated by 8% polyacrylamide gel electrophoresis to obtain genotyping. The primer sequence is shown in Supplementary Table S1.
Candidate gene sequencing analysis
The complete coding sequence (CDS) of candidate genes was cloned and sequenced based on the reference genome (ZAAS_Lsic_2.0) (Supplementary Table S1). Total RNA extraction using the Eastep Super Total RNA Extraction Kit (Promega, Shanghai, China) according to manufacturer ‘s instructions. The first strand cDNA was synthesized using reverse transcriptase 5× All-In-One RT Master Mix (abm, Mississauga, Ontario, Canada). 2 × Phanta Max Master Mix (Dye Plus) PCR Premix (Vazyme, Nanjing, China) for PCR amplification. The PCR product was detected by 1.0% agarose gel electrophoresis, and the target product was purified and recovered by gel cutting. The expression vector was constructed following the manufacturer’s instructions using the CV16-Zero Background pTOPO-Blunt Cloning Kit from Aidlab (Beijing, China). The vector was transformed into Trans5α chemically compatible cells according to the manufacturer’s instructions (TransGen Biotech, Beijing, China), and monoclonal colonies were picked for sequencing. DNA fragment sequencing completed by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). DNA sequencing results were aligned using DNAMAN v.9 software (Lynnon Biosoft, CA, United States).
RACE-based cloning of full-length cDNAs of candidate genes
Rapid amplification of cDNA 5’ ends (5’ RACE) was performed using the 5’-RACE Kit (Sangon, Shanghai), according to the manufacturer’s instructions. Specific primers were designed based on the validated cDNA sequences (Supplementary Table S1). Total RNA was extracted using the Eastep Super Total RNA Kit (Promega, Shanghai, China). Using a specific sequence in the mRNA as a binding site, the first strand cDNA was synthesized with the sequence-specific reverse transcription primer RT1/RT2 and the reagent, Reverse Transcriptase Mix (RNase H). After annealing with TdT enzyme plus 10-15 (dC) residues, (dC) residues were paired with a 5 ‘ universal adaptor primer containing the oligonucleotide sequence to perform the first round of PCR amplification using sequence-specific primer R1 as a downstream primer and first strand cDNA as template. A universal 5’ RACE outer primer containing a partial splice sequence was then used as an upstream primer. Using another specific primer R2 as a downstream primer, a cDNA fragment from the 5’ end of the gene of interest was amplified. The amplified PCR products were cut and purified by agarose gel electrophoresis, and the target products were recovered for cloning and sequencing.
Rapid amplification of cDNA 3’ ends (3’ RACE) was performed using the 3’-RACE Kit (Sangon, Shanghai), according to the manufacturer’s instructions. Specific primers were designed based on the validated cDNA sequences (Supplementary Table S1). First-strand cDNA was synthesized by reverse transcription using the poly (a) tail at the 3’ end of the mRNA as the 3’ adaptor for the primer binding site. Specific primer 3RF1 was used as the upstream universal primer for 3’ RACE. An 3’ RACE Outer Primer containing a partial splice sequence was used as a downstream primer. The first round of PCR amplification was performed using the first strand of cDNA as a template. The specific primer 3RF2 was then used as an upstream primer. Further, a universal primer containing a partial linker sequence, the 3’ race inner primer, was used as a downstream primer. The second round of PCR amplification was performed to amplify the DNA fragment at the 3’ end of the target gene. The amplified PCR products were purified by agarose gel electrophoresis, and the target products were recovered for cloning and sequencing. The complete sequence of the target gene was obtained by assembly and splicing of the sequence.
RNA extraction and qRT-PCR analysis of candidate genes
To understand the expression pattern of candidate genes in different spatial and temporal contexts, quantitative real-time PCR (qRT-PCR) was performed during different stages of fruit development and in different plant organs. Total RNA was extracted from the pericarp at different stages of development and from male flowers, roots, stems, and leaves at the flowering stage. cDNA was synthesized using the Reverse Transcriptase RT Master Mix (Takara, Japan). SYBR Green real-time fluorescent quantitative PCR mixture was used for all the reactions (Supplementary Table S1). qRT-PCR analysis was performed using a QTOWER 2.2 qPCR instrument (Jena, Germany). The reactions mixture was prepared with 2 μL of cDNA (50 ng/μL), 0.8 μL of forward primer (10 μm), 0.8 μL of reverse primer (10 μm), 0.4 μL of ROX Reference Dye II (50×), 10 μL TB Green Premix Ex Taq II (Tli RNaseH Plus) (2×), and 6 μL of sterilized distilled water, and the final volume was made up to 20 μL followed by instant centrifugation. Preheating at 95°C for 30 s, followed by heating at 95°C for 5 s and at 60°C for 34 s for a total of 40 cycles, high-resolution melting at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Three replicates were performed for each sample. Relative expression was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Subcellular localization of LsAPRR2
Primer design based on the complete coding sequence (CDS) of HG_GLEAN_10010973 was obtained by RACE (Supplementary Table S1). The target fragment without the stop codon was amplified by PCR and cloned into the modified vector pBI221-EGFP (Li et al., 2012; Li et al., 2022). The 35S::GFP and 35S::GFP-LsAPRR2 fusion vectors were transformed into Agrobacterium tumefaciens GV3101 strain. Transformed Agrobacterium tumefaciens cells were infiltrated into the inner epidermal cells of onion bulbs and incubated at 25°C in a light incubator. GFP signals were detected at 16h post-transformation using a laser confocal scanning microscope LEICA-TCS-SP8MP (Leica Germany), and nuclei were stained using 4 ‘, 6-diamidino-2-phenylindole (DAPI). The excitation wavelength was 488 nm.
LsAPRR2 promoter cloning and GUS activity analysis
The first 2000 bp of the biparental LsAPRR2 promoter were cloned (Supplementary Table S1) and subsequently ligated into an expression vector (pBI121) containing a β-Glucuronidase (GUS) reporter gene (Li et al., 2022). The Agrobacterium tumefaciens GV3101 strain was transformed with the fusion reporter vectors as well as the empty vector. Transformed Agrobacterium cell suspension was injected into tobacco leaves and samples were immersed in X-Gluc buffer 7 days after injection (12 mM potassium ferricyanide, 0.3% (v/v) Triton X-100, 12 mM potassium ferrocyanide and 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-glucuronide). The buffer was allowed to penetrate the samples under vacuum, stained overnight at 37°C, decolorized by several washes in 75% (v/v) ethanol, and photographed after complete decolorization of the chlorophyll (Koo et al., 2007).
Phylogenetic analysis
To elucidate the relationship between the LsAPRR2 protein and its homologs, we performed a phylogenetic analysis of the LsAPRR2 homologs. The APRR2 protein sequences of some Cucurbitaceae, Solanaceae and Brassicaceae species were downloaded from NCBI (NCBI, Bethesda, MD, USA). Phylogenetic trees with 1000 Bootstrap repeats were constructed by Neighbor-joining method of MEGA 6.0 (Mega Limited, Auckland, New Zealand) software.
Close-linked molecular marker development
Marker development was done via Primer Premier 5 software (Premier, Canada) (Supplementary Table S1). The total volume of PCR reaction was 12.0 μL, with 2.0 μL DNA (20-50 ng/μL), 1.0 μL each of forward and reverse primers (10 μm), 6.0 μL 2×PCR Master Mix, and 2.0 μL ddH2O. The reaction conditions were as follows: heat treatment at 95°C for 3 min, denaturation at 95°C for 30 s, annealing at 55-60°C for 30 s, extension at 72°C for 30~35 cycles; complete extension at 72°C for 5min; cool down to 4°C for storage. PCR amplicons were separated by electrophoresis using 8% native polyacrylamide gels with a voltage of 240 V and a current of 300 mA, and the program was run for 2 h. Fifty-one bottle gourd germplasm resources were validated. These germplasm resources were divided into two populations of green and white-colored fruits.
Results
Genetic and phenotypic analysis of epidermal color in bottle gourd
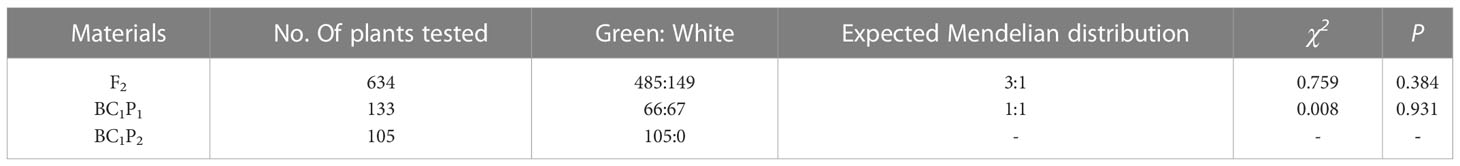
To provide an accurate description of the changes in epidermal pigmentation pattern in bottle gourd during fruit development, we plotted chlorophyll and carotenoid profiles with growth period (Figures 1C, D). In the inbred combination H06×H16, the paternal line H06 had white pericarp, the maternal line H16 had green pericarp, and all F1 pericarp had green pericarp. Trait segregation in pericarp color occurring in the F2 population unraveled 149 white phenotypes and 485 green phenotypes, with a separation ratio of 1:3, respectively (χ2 = 0.759, ρ = 0.384). The BC1P1 plants were segregated as 67 white individuals and 66 green individuals, a distribution consistent with a 1:1 skew analysis ratio (χ2 = 0.008, ρ = 0.931). All the individuals harvested for BC1P2 had green epidermis (Table 1).
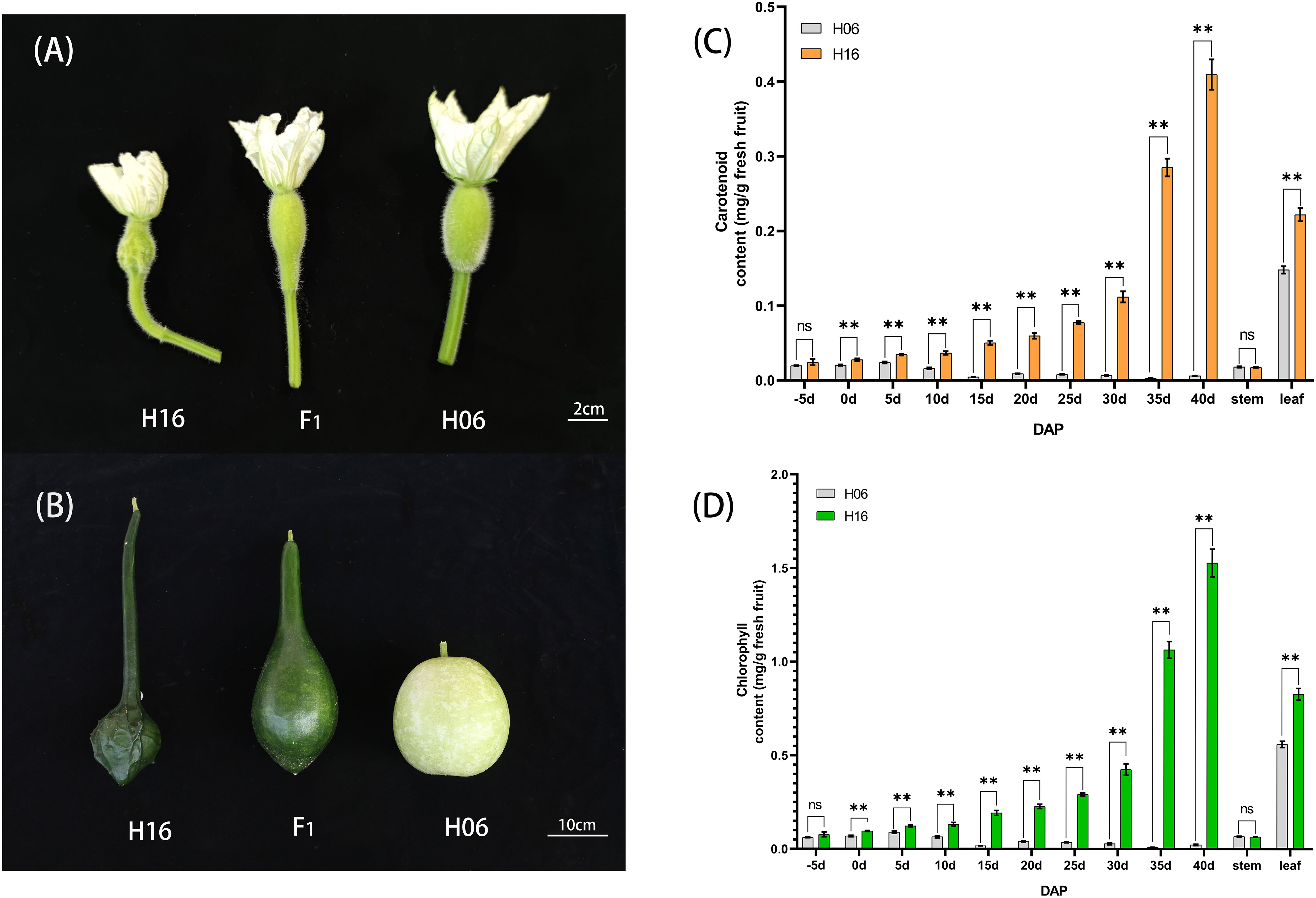
Figure 1 Phenotypic characteristics and pigment content analysis of gourd rind at different growth stages. (A) Phenotype on the day of pollination in H16, F1 and H06; Bar=2cm. (B) Phenotypes 30 days after pollination in H16, F1 and H06; Bar=10cm. (C, D) Pericarp Pigment Contents, and stem, leaf pigment contents from 5 days before pollination to 40 days after pollination for both parents. (C) Carotenoid content. (D) Chlorophyll content. *, 0.01<P<0.05; **, P<0.01. ns indicates no statistical difference.
Chlorophyll content determination and chloroplast observation
To study the changes in pericarp pigment content during fruit development, we determined the chlorophyll and carotenoid content of the pericarp of two parents at different stages of fruit development (-5, 0, 5, 10, 15, 20, 25, 30, 35, 40DAP). The chlorophyll content of the green-skinned parent (H16) was higher than that of the white-skinned parent (H06) at all tested stages (Figure 1D). The chlorophyll content of green pericarp (H16) increased exponentially with fruit growth, while that of the white pericarp (H06) experienced a brief trace increase after pollination and then decreased to a lower level. Chlorophyll content measurements of both the parents’ pericarp were correlated with the visual inspection. In addition, the carotenoid content trend in both parents’ pericarp was consistent with that of chlorophyll (Figure 1C). The above results suggested that the H16 pericarp accumulated more pigments during growth, which prompted us to clarify further the differences in chloroplasts in H16 and H06 cells.
The bi-parental fruits at 30 DAP were harvested for light microscopic observation of pericarp cells. The results showed no significant difference in the size of mature cells between the two parents. In the white parent (H06), almost no chloroplasts were observed in the pericarp cells, with only visible chloroplast structures around the stomata, and the peel color was pale yellow. In contrast, in the green parent (H16), many chloroplast structures were observed in the pericarp cells, with larger and fuller chloroplasts with a dark green peel color (Figure 2).
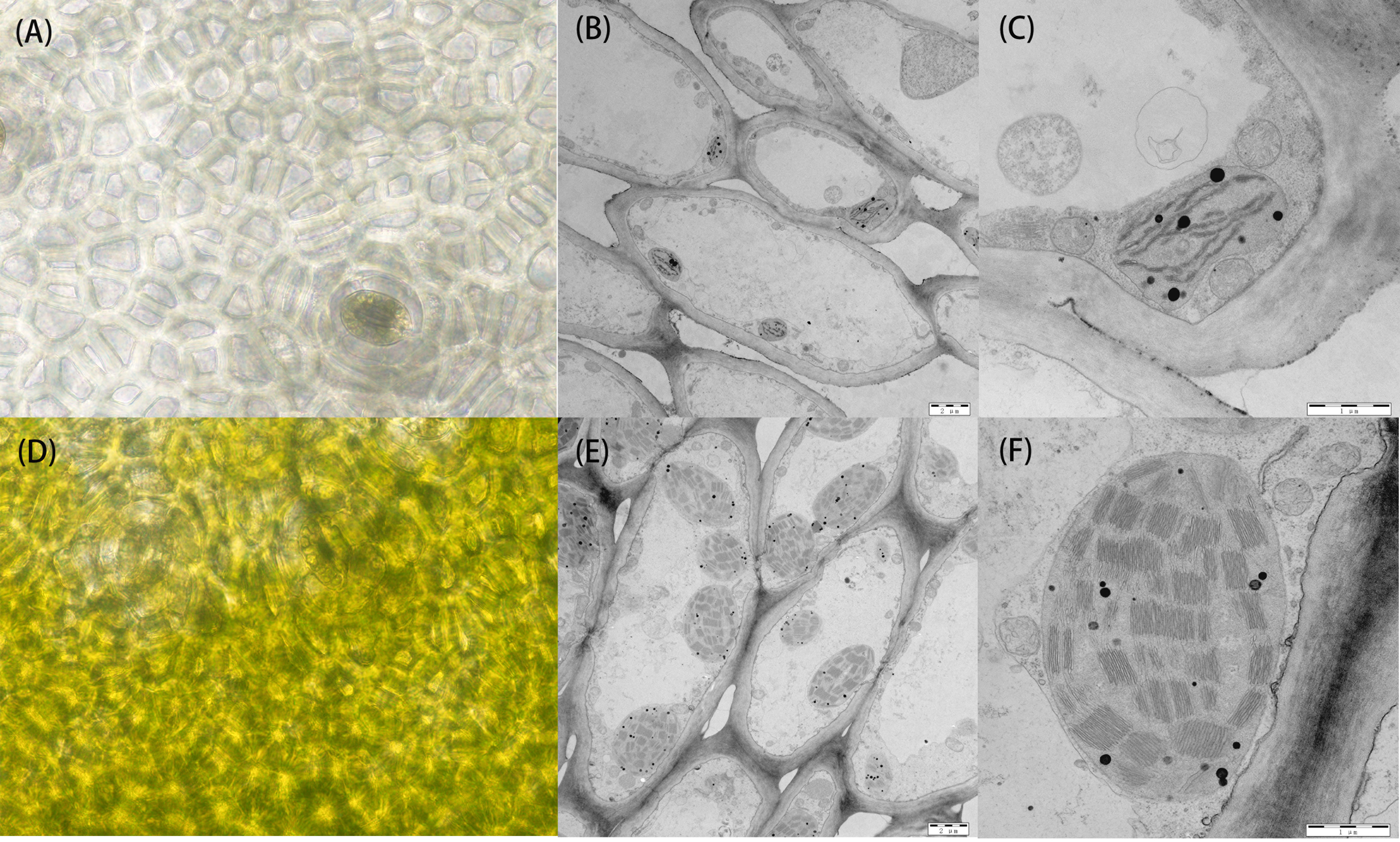
Figure 2 Cytology and transmission electron microscopy of 30 DAP peel from both parents. (A) Cytological observation of the white parental H06 pericarp. (B, C) transmission electron microscopy observations of H06 cells and chloroplasts, respectively. (D) Cytological observation of green parental H16 pericarp. (E, F) transmission electron microscopy observations of H16 cells and chloroplasts, respectively.
To research the structural changes in chloroplasts, we used transmission electron microscopy (TEM) to analyze amphiphilic epidermal cells’ chloroplasts. It was concluded that the white parent (H06) had a low number of intracellular chloroplasts, a small chloroplast volume, and an atrophic thylakoid structure in the epidermal cells. In contrast, the green parental (H16) epidermal cell chloroplasts were more numerous, larger in size with densely packed thylakoid stacks, and plump. The chloroplasts occupied much of the cell space in the green parental epidermal cells (Figure 2).
Mapping of candidate genes to the first end of chromosome 1 using BSA-seq
BSA-seq-based resequencing yielded about 32,591,680 and 30,922,232 clean reads from H06 and H16 parents, respectively. In addition, 34,971,621 and 34,218,279 clean reads were obtained from the green and white mixer pools. The mean Q30 value reached 92.53 with a GC content range of 32.06 to 32.88%. The average matching efficiency of the samples to the reference genome (ZAAS_Lsic_2.0) was 99.34%, with an average coverage depth of 26.75×. The genome coverage of 99.28% (at least one base covered) was obtained in this study. Quality control indicated successful sequencing, and the data was used for subsequent mutation detection and correlation analysis. A total of 657,869 single nucleotide polymorphisms (SNPs) were detected in the four gene banks, of which 48,241 were high-quality SNPs used to calculate the SNP index between two F2 populations. Preliminary BSA-seq localization results were obtained with the ED and ΔSNP-index association algorithms. A confidence interval (99%) associated with the peel color trait was identified at the top end of chr01, spanning a 5.25 Mb region from 0 Mb to 5.25 Mb from the start of chr01 (Figure 3A). This region contained 467 genes, and SNP analysis revealed that 116 of these genes had nonsynonymous mutations.
Figure 3 Fine mapping of the LsAPRR2 gene in bottle gourd. (A) Distribution map of Euclidean distance (ED) association values on chromosomes for fruit coat color traits. (B) The candidate gene LsAPRR2 is located within a 22,645bp interval between the flanking markers Sp2.1 and Sp2.2. (C) Genotyping of recombinant plants from F2, with plant numbers and phenotypic information listed on the right. (D) Schematic diagram of the LsAPRR2 gene structure, with white boxes indicating the 5’UTR and 3’UTR positions of the gene and black rectangles and solid lines representing the positions where exons and introns are located, respectively.
Fine mapping of LsAPRR2
To further narrow the candidate region, five pairs of KASP markers were developed at an average region of 1 Mb within the target region of 5.25 Mb initially targeted by BSA-seq. Five KASP markers were used for the combined genotype-phenotype analysis of 634 F2 individuals. The candidate region was between markers Sp2 and Sp3, and the region contained a physical distance of 0.94 Mb (Figure 3B). To further narrow the candidate region, five polymorphic InDel Markers between Sp2 and Sp3 were developed for the combined genotype-phenotype analysis of 64 recombinant plants from this region. The candidate region was mapped between markers Sp2.1 and Sp2.3, with an region physical distance of 121.37 Kb containing 11 genes. A population comprising 5208 F2 isolates was used to further narrow the region, which ultimately anchored the region between markers Sp2.1 and Sp2.2 at a physical distance of 22.645 Kb (Figure 3B). Gene annotation revealed that two annotated genes (HG_GLEAN_10010972, HG_GLEAN_10010973) were located within this spacer.
RACE technique proved that LsAPRR2 was the only gene in the interval
A partial cDNA sequence of HG_GLEAN_10010973 was obtained from the bottle gourd reference genome (ZAAS_Lsic_2.0). Using this sequence fragment as a template, the complete sequence of HG_GLEAN_10010973 was amplified by 5’ RACE and 3’ RACE, where the amplicon lengths were 240 bp and 68 bp, respectively. The full-length cDNA sequence of the gene was obtained by splicing three fragments and deleting the duplicate fragments. The protein sequence encoded by the cDNA of this gene consisted of 514 amino acids. After mapping this cDNA to the genome, we found that the 5’UTR of the HG_GLEAN_10010973 gene contained HG_GLEAN_10010972. Therefore, we concluded that the gene HG_GLEAN_10010972 is essentially a part of the HG_GLEAN_10010973 gene, and the 5’ UTR sequence of HG_GLEAN_10010973 was incorrectly annotated as a separate gene during genome assembly-based prediction. These observations highlighted that the markers Sp2.1 and Sp2.2 intervals contained only one gene, HG_GLEAN_10010973, which was named LsAPRR2 in the current study (Figure 3).
Sequence alignment and expression analysis of LsAPRR2
To analyze the sequence of LsAPRR2, we sequenced the full-length CDS of LsAPRR2, cloned from both the parents, and compared the sequencing results using DNAMAN v.9. The full-length CDS of LsAPRR2 was 1542 bp and consisted of 12 exons. In contrast to H16, two nonsynonymous nucleotide mutations (A→G) (G→C) were detected in the CDS sequence of H06, which caused two amino acid substitutions in the H06 CDS sequence (I→M) (G→A). We used SMART (https://smart.embl.de/) for LsAPRR2 protein prediction, which showed that the protein structure of LsAPRR2 contains two important conserved functional domains, the REC domain (18-128 bp) and the Pfam: Myb_DNA-binding domain (279-329 bp). The two altered amino acids in the predicted results for the white epidermal parent were located in the non-conserved structural domain of the LsAPRR2 protein.
Spatiotemporal expression analysis of LsAPRR2 in both parents was performed using qRT-PCR. Differential expression analysis of the pericarp (-5, 0, 5, 10, 15, 20, 25, 30, 35, 40 DAP) and different tissues (root, stem, leaf, flower) showed that expression of LsAPRR2 was significantly higher in the pericarp of H16 than that of H06 at different times. The difference in expression reached a maximum at 30 DAP, with H16 expression reaching 133.6 times that of H06, followed by a decline in its expression. Lower LsAPRR2 expression was observed during H06 fruit development throughout the period. The expression of LsAPRR2 was higher in flowers than in roots, stems, and leaves (Figure 4).
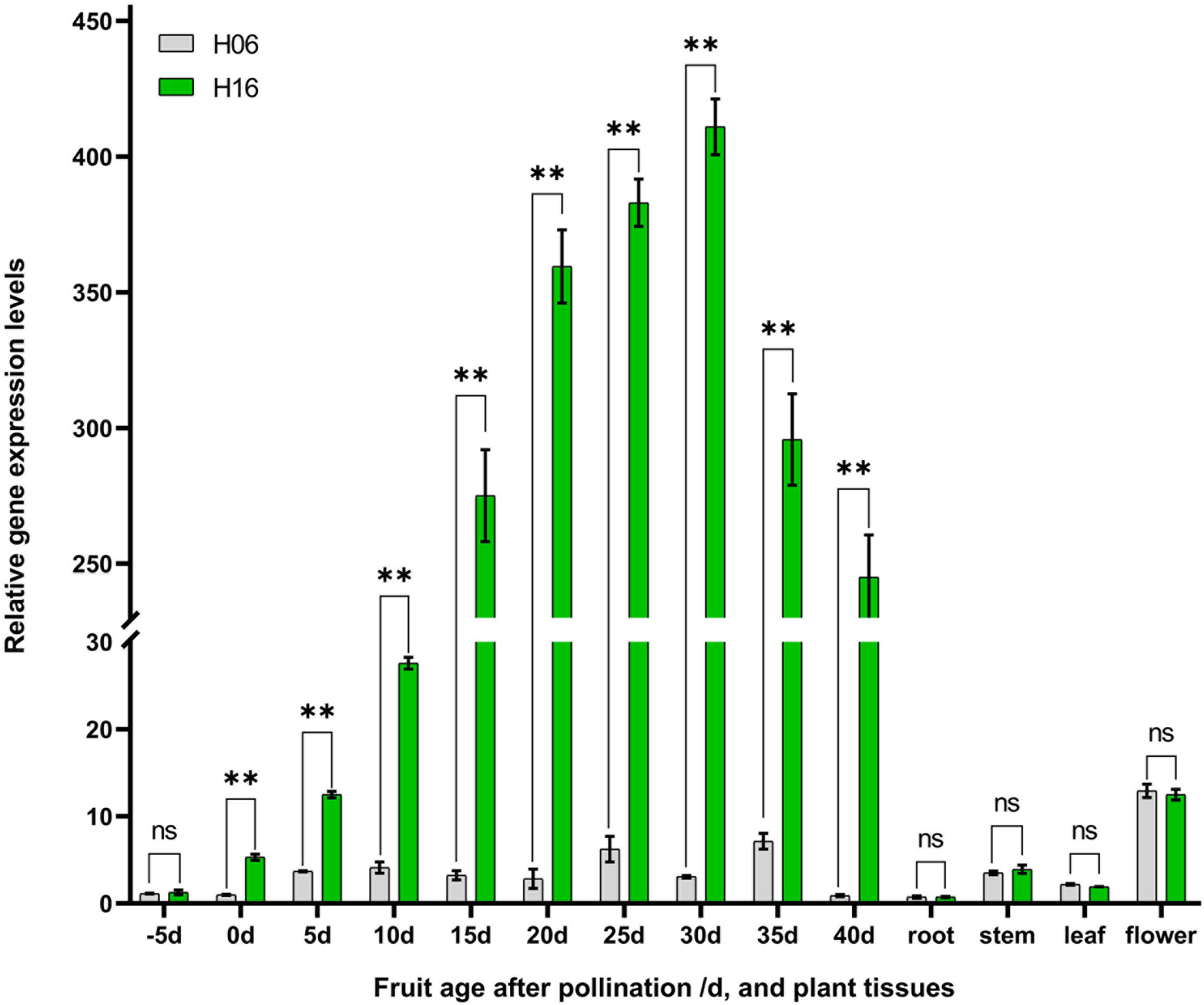
Figure 4 Analysis of LsAPRR2 expression. Differential analysis of LsAPRR2 expression in fruit epidermis from 5 days before pollination to 40 days after pollination in H06 and H16, and in root, stem, leaf, male flower. *, 0.01<P<0.05; **, P<0.01. ns indicates no statistical difference.
Subcellular localization of LsAPRR2
To determine the subcellular location of the LsAPRR2 protein, 35S::GFP and 35S::GFP-LsAPRR2 were transformed into onion inner epidermal cells. As shown in Figure 5, the empty vector 35S::GFP fluorescent signal was observed in whole onion cells, while the 35S::GFP-LsAPRR2 fusion vector expression was only observed in the nuclei. The blue fluorescent signal for the DAPI stain was observed in all cell nuclei, which overlapped with the expression of the 35S::GFP-LsAPRR2 fusion vector.
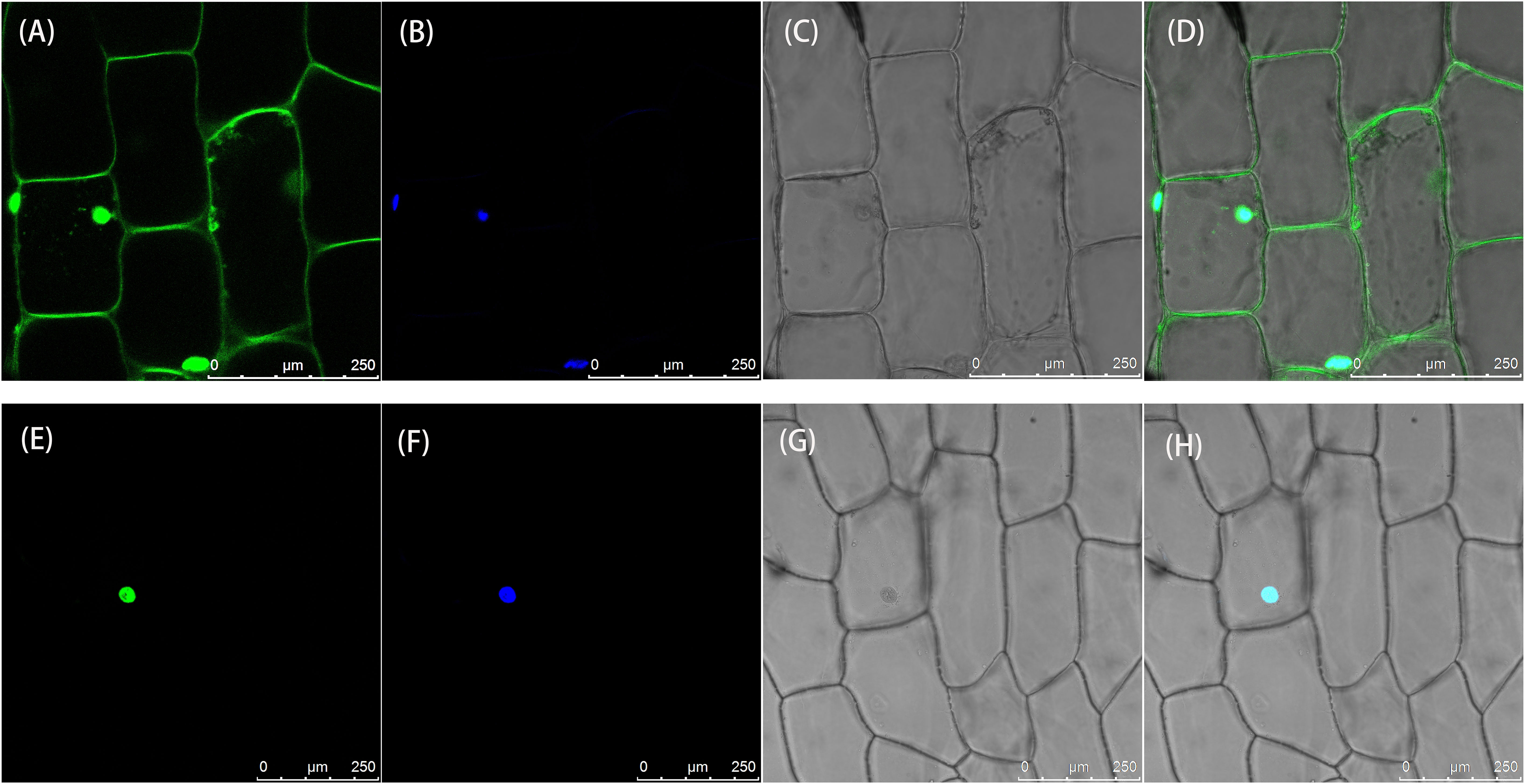
Figure 5 Subcellular localization of LsAPRR2. (A–D) Subcellular localization of the empty vector 35S::GFP. (E–H) Subcellular localization of 35S::GFP-LsAPRR2. (A) and (E) are the GFP fluorescence channels (green color). (B) and (F) are the DAPI fluorescence channels (glue color). (C) and (G) are bright field. (D) and (H) are overlay images.
LsAPRR2 promoter cloning and GUS activity analysis
We found that the expression of LsAPRR2 in the rind of green-skinned bottle gourd (H16) and white-skinned bottle gourd (H06) differed significantly. To clarify all unknown variants between the two parents, we performed cloning and sequence analysis of the 2000 bp upstream region of the start codon. Sequence alignment unraveled a large number of mutations within the -991~-1033 bp promoter region upstream of the start codon (ATG). The white parent had 11 base insertions and 8 SNP mutations in this region compared to the green parent. We used PlantCARE(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)for the prediction of cis-acting elements within this region and the results showed that variants in this region had altered binding sites for TATA-box promoter elements. It has been shown that the TATA-box binding site is the core promoter element at the transcription start site around -30 bp, and alteration in this site affects the strength of the promoter (Jores et al., 2021). The GUS reporter assay showed that the 35S: GUS fusion vector had the strongest expression activity, followed by the LsAPRR2Green: GUS fusion vector. The LsAPRR2White: GUS fusion vector showed almost no expression (Figure 6). This experiment demonstrated that variants in the promoter -991~-1033 bp region affect the expression of the LsAPRR2 gene.
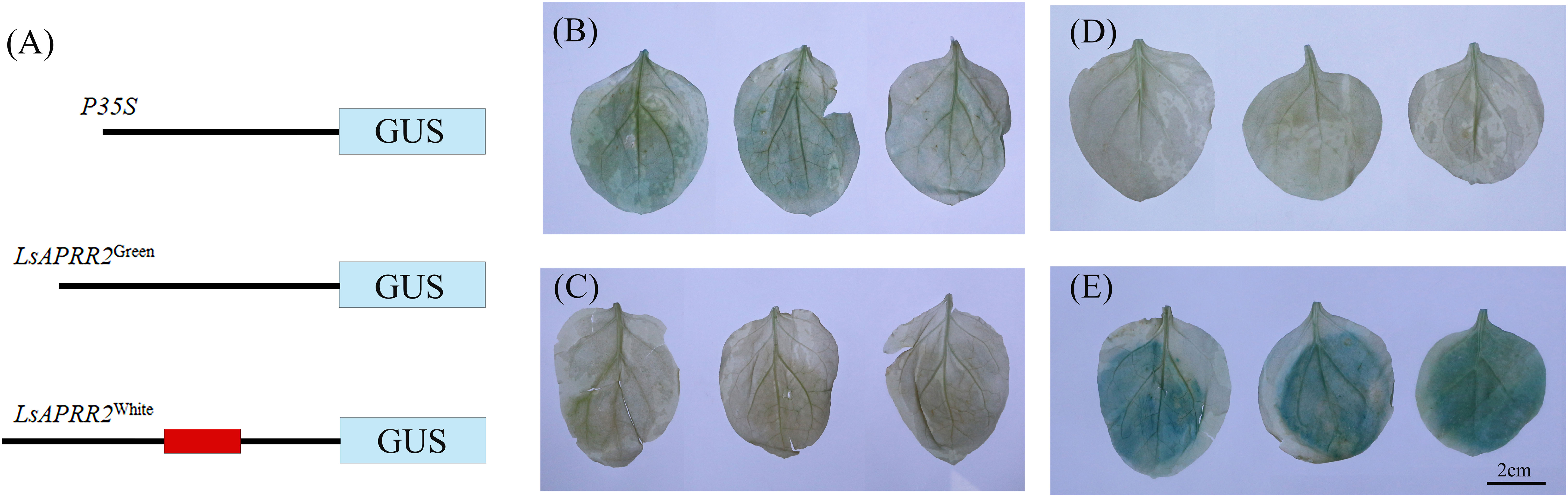
Figure 6 Schematic diagram of GUS fusion vector and histochemical determination of GUS activity. (A) Schematic diagram of the LsAPRR2Green and LsAPRR2White promoter-β-Glucuronidase (GUS) fusion vectors. (B–E) Histochemical assay of GUS activity in leaves of Nicotiana benthamiana (B) LsAPRR2Green:GUS. (C) LsAPRR2White : GUS. (D) CK. (E) P35S:GUS.
Phylogenetic analysis
We used a BLAST search in the NCBI database to obtain the sequence information, followed by phylogenetic analysis to elucidate the relationship between the LsAPRR2 protein and its homologs from different plant species (Benincasa hispida, Melon, Cucumis sativus, Cucurbita moschata, Cucurbita pepo, Bitter gourd, Solanum lycopersicum, Capsicum annuum, Nicotiana benthamiana, Solanum melongena L.) (Figure 7). The phylogenetic tree indicated that (LsAPRR2) was closely related to the Cucurbitaceae crops (Benincasa hispida, Melon, Cucumis sativus, Cucurbita moschata).

Figure 7 Phylogenetic analysis. Evolutionary relationship between LsAPRR2 and its cognate APRR2 protein.
Development of molecular breeding aided markers
Using the sequences targeting the deletion variants in the upstream promoter of LsAPRR2, we developed an InDel molecular breeding-assisted marker G0913 that resulted in 362 bp and 351 bp target fragments from H06 and H16 respectively. We validated the marker against 133 BC1P1 and 5842 F2 populations and recorded the phenotypes and genotypes in both populations. The result showed the phenotype and genotype were 100% matche. Further, we verified 49 selfed lines of material using the G0913 marker, and the results showed that 46 of these 49 selfed lines were genotypically consistent with their phenotypes, with a concordance rate of 93.88% (Figure 8).
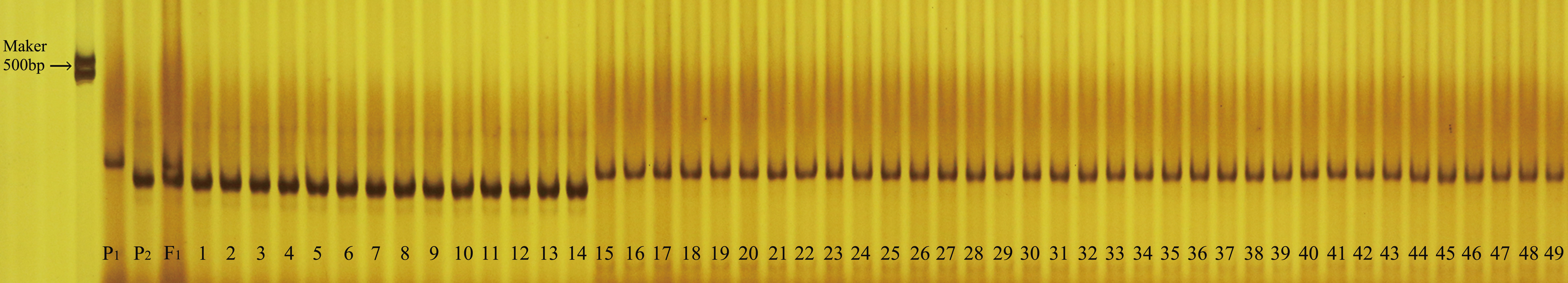
Figure 8 Validation of molecular breeding aid markers in 49 bottle gourd self-crossing populations. P1 is the white parent H06, P2 is the green parent H16, F1 is a P1, P2 cross. Numbers 1-17 are green-skinned gourds, numbers 18-49 are white-skinned gourds.
Discussion
Peel color is one of the most important phenological qualities of bottle gourds, and thus, it is a trait of special interest for breeders. In the current study, we observed that green skin was more dominant than white skin. Further, genetic analysis of the bottle gourd peel color trait in a six-generation genetic population proved that the green peel color of bottle gourd is inherited as a single dominant gene. This is consistent with genetic models of peel color in other Cucurbitaceae crops, such as cucumber (Liu et al., 2016), wax gourd (Ma et al., 2021), and watermelon (Li et al., 2019). However, the genes that specifically regulate the peel color trait are still unknown in bottle gourd.
With the development of high-throughput sequencing technology, whole genome resequencing has become cost-effective, and data access has been more accurate and efficient. We used BSA-seq sequencing for rapid and accurate interval targeting of peel color trait-associated genes in our study. Anchoring of genes to a 1,085,238-1,107,883 bp region on chromosome 1 was completed using a novel single nucleotide polymorphism typing technique (KASP) in combination with conventional InDel markers (Figure 3), and the full length of the HG_GLEAN_10010973 gene was cloned using RACE. We demonstrated that the HG_GLEAN_10010972 gene within the candidate interval is a part of the HG_GLEAN_10010973 5’UTR, highlighting a previous genomic prediction error. The final interval contained only one gene, HG_GLEAN_10010973, which was annotated as a two-component response regulator-like APRR2 isoform ×4. We found that APRR2-Like was its homolog in Arabidopsis. Therefore the gene HG_GLEAN_10010973 was named LsAPRR2 in our study. Ou (2019) reported that the gene MELO3C003375, controls peel color in melon. In the melon fruits with the non-green epidermis, a 13 bp insertion was identified in the MELO3C003375 gene, which altered the epidermal coloration of the melon. The candidate gene APRR2 affecting green pericarp was identified in cucumber, and a single base insertion was discovered in the white pericarp gene w. The insertion resulted in the occurrence of a premature stop codon that disables the allele involved in chlorophyll accumulation and chloroplast development (Liu et al., 2016). A candidate gene Bch05G003950 (BhAPRR2) controls peel color in wax gourd. A two-base (GA) deletion was found in the coding sequence of BhAPRR2 in white rind winter melon resulting in a premature stop codon, affecting the peel color (Ma et al., 2021). Elad et al. (2019) found that an AG→AC mutation at the junction of intron 6 and exon 7 in the CLAPRR2 gene, which controls watermelon peel color, resulted in a variable cut in this region, causing a 16 bp deletion in the coding region of light green striped watermelon that triggered an early termination of the transcript. Our study identified two nonsynonymous nucleotide mutations in H06 compared to H16 (A→G) (G→C). These two nucleotide variants caused a substitution of two amino acids in the protein sequence encoded by H06 compared to H16 (I→M) (G→A). Further, cloning and sequence comparison of the two parental LsAPRR2 promoter regions revealed eleven base insertions and eight SNP mutations in the -991 to -1033 bp region in the white pericarp parent (H06). GUS assay indicated that mutations in the promoter region alter the expression of the LsAPRR2 gene (Figure 6). LsAPRR2Green : GUS expression intensity was significantly higher than that of LsAPRR2White : GUS. In recent years, the APRR2-Like gene has been shown as a key transcription factor involved in pericarp chloroplast regulation in a variety of crops (Powell et al., 2012; Pan et al., 2013; Liu et al., 2016; Elad et al., 2019; Ma et al., 2021).
In our study, we found that the nuclear gene LsAPRR2 influenced the color of gourd rind by regulating the development of chloroplasts. Microscopic observation of the pericarp cells showed no significant difference in size between the two parents (Figure 2), unlike the results described by Wang et al. (2020), where green pericarp cells were found to be smaller than white pericarp cells. The difference in pericarp cell size might be due to differences in the experimental material. Transmission electron microscopy observed that chloroplasts present in the epidermal cells of green bottle gourd were several times larger in volume than those in white epidermal cells (Figure 2). These findings were inconsistent with Liu et al. (2016), where the number of chloroplasts mainly caused the difference in coat color between cucumber green parent and white parent, but similar to the results by Zhu et al., 2022, in which chloroplast volume mainly affected chlorophyll accumulation leading to the changes in color. In a spatiotemporal expression analysis of LsAPRR2 (Figure 4), the expression of LsAPRR2 in the green pericarp parent (H16) was upregulated at the early stages of fruit development and gradually decreased with fruit growth after reaching 30 DAP. The white pericarp parent (H06) exhibited a small increase in LsAPRR2 expression a few days before pollination and then remained at a low expression level. LsAPRR2 expression in roots, stems, and leaves of the two parents was low and not significantly different (Figure 4), which is consistent with the pattern of variation in APRR2 expression in fruit epidermis that has been reported in cucumber and wax gourd (Liu et al., 2016; Ma et al., 2021). Based on the chlorophyll measurement curves, we concluded that the chlorophyll content of H16 pericarp was exponentially increased after pollination and was still on the rise when APRR2 expression was downregulated after 30 DAP (Figure 1). These findings were inconsistent with the studies in tomato (Pan et al., 2013) and cucumber (Liu et al., 2016), where APRR2 expression was gradually upregulated after pollination and then decreased. Studies have shown that the gradual degradation of fruit chlorophyll during developmental stages is caused by a negative balance between synthesis and degradation of chlorophyll (Jacob-Wilk et al., 1999). However, green-skinned bottle gourds do not undergo a de-greening phase during growth. Rapid degradation of chlorophyll is a characteristic of green organs during senescence or maturation (Zhu et al., 2015). Therefore, we speculated that bottle gourd might have a different pattern of chlorophyll degradation regulation than tomato and cucumber, which undergo a de-greening phase during fruit development. We measured high chlorophyll content, whereas low expression of APRR2 in leaves (Figures 1, 4). Substantial pieces of evidence suggest that APRR2 may be a chlorophyll regulator specifically expressed in the pericarp (Powell et al., 2012; Pan et al., 2013; Nadakuduti et al., 2014; Ma et al., 2021);. However, some recent studies have shown that APRR2 may also be involved in color regulation in non-fruiting organs of different plants (Zhu et al., 2022) and may perform some non-color-regulated roles (Jiang et al., 2022).
Molecular marker-assisted breeding can significantly fasten the breeding process (Nadeem et al., 2018). Currently, several markers applicable in molecular marker-assisted breeding have been developed in some Cucurbitaceae crops, such as cucumber (Liu et al., 2015) and winter melon (Ma et al., 2021). We have developed an InDel molecular breeding-aided marker for a deletion variant in the upstream promoter sequence of the target gene LsAPRR2 (Figure 8), which can distinguish between green and white bottle gourds (Accuracy rate of 93.88%). The lack of 100% accuracy is that there may be other genes involved in the regulation of the green epidermis of the gourd, which reduces the accuracy of the molecular markers. Based on our findings, selfed lines can be identified in the future with no variation in the LsAPRR2 gene but with phenotypic variation to pinpoint other genes that regulate bottle gourd peel color. Also, a system of molecular markers can be generated subsequently associated with bottle gourd peel color to achieve accurate breeding for this trait.
Data availability statement
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0004006. https://db.cngb.org/search/?q=CNP0004006.
Author contributions
YY and PW conceived and designed the experiments. YH, GZ, MS performed the experiments. GZ, MS, RZ, SH, XZ collected some data. YH analyzed the data. YH wrote the manuscript. YY, WY, ZL, PW revised the manuscript. YH, YY and PW contributed equally. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Modern Agricultural Industry Technology System Guangxi Vegetables and Fruits Innovation Team Cultivation and Breeding Function Expert Project (20181235). The study was also supported by the Guangxi Innovation-Driven Development Special Fund Major Project (Guike AA17204039-2) and the Guangxi Innovation-Driven Development Special Fund Major Project (Guike AA17204026-1).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1130669/full#supplementary-material
References
Azuma, A., Kobayashi, S., Mitani, N., Shiraishi, M., Yamada, M., Ueno, T., et al. (2008). Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor. Appl. Genet. 117, 1009–1019. doi: 10.1007/s00122-008-0840-1
Brand, A., Borovsky, Y., Hill, T., Rahman, K., Bellalou, A., Van, D., et al. (2014). CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 127, 2139–2148. doi: 10.1007/s00122-014-2367-y
Dong, S. Y., Miao, H., Zhang, S. P., Liu, M. M., Wang, Y., Gu, X. F. (2012). Genetic analysis and gene mapping of white fruit skin in cucumber (Cucumis sativus l). Acta Botanica Boreali-Occidentalia Sin. 32 (11), 2177–2181.
Elad, O., Galil, T., Vexler., L., Asaf, D., Ayala, M., Adi, F., et al. (2019). The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. J. Exp. Bot. 70, 3781–3794. doi: 10.1093/jxb/erz182
Erickson, D. L., Smith, B. D., Clarke, A. C., Sandweiss, D. H., Tuross, N. (2005). An Asian origin for a 10,000-year-old domesticated plant in the americas. P Natl. Acad. Sci. U.S.A. 102, 18315–18320. doi: 10.1073/pnas.0509279102
Feder, A., Burger, J., Gao, S., Lewinsohn, E., Katzir, N., Schaffer, A. A., et al. (2015). A kelch domain-containing f-box coding gene negatively regulates flavonoid accumulation in muskmelon. Plant Physiol. 169, 1714–1726. doi: 10.1104/pp.15.01008
Fitter, D. W., Martin, D. J., Copley, M. J., Scotland, R. W., Langdale, J. A. (2002). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31, 713–727. doi: 10.1046/j.1365-313X.2002.01390.x
Hao, N., Du, Y. L., Li, H. Y., Wang, C., Wang, C., Gong, S. Y., et al. (2018). CsMYB36 is involved in the formation of yellow green peel in cucumber (Cucumis sativus l.). Theor. Appl. Genet. 131, 1659–1669. doi: 10.1007/s00122-018-3105-7
He, C., Holme, J., Anthony, J. (2014). SNP genotyping: the KASP assay. Methods Mol. Biol. 1145, 75–86. doi: 10.1007/978-1-4939-0446-4_7
Hu, Z. H., Xu, F., Guan, L. P., Qian, P. P., Liu, Y. Q., Zhang, H. F., et al. (2014). The tetratricopeptide repeat-containing protein slow green1 is required for chloroplast development in arabidopsis. J. Exp. Bot. 65, 1111–1123. doi: 10.1093/jxb/ert463
Jacob-Wilk, D., Holland, D., Goldschmidt, E. E., Riov, J., Eyal, Y. (1999). Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated citrus fruit and its regulation during development. Plant J. 20, 653–661. doi: 10.1046/j.1365-313X.1999.00637.x
Jia, T., Cheng, Y. T., Khan, I., Zhao, X., Gu, T. Y., Hu, X. Y. (2020). Progress on understanding transcriptional regulation of chloroplast development in fleshy fruit. Int. J. OF Mol. Sci. 21, 6951. doi: 10.3390/ijms21186951
Jiang, L., Fu, Y. X., Tian, X. K., Ma, Y. H., Chen, F. D., Wang, G. D. (2022). The Anthurium APRR2-like gene promotes photosynthetic pigment accumulation in response to salt stress. Trop. Plant Biol. 15, 12–21. doi: 10.1007/s12042-021-09305-3
Jores, T., Tonnies, J., Wrightsman, T., Buckler, E. S., Cuperus, J. T., Fields, S., et al. (2021). Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters. Nat. Plants 7, 842. doi: 10.1038/s41477-021-00932-y
Koo, J., Kim, Y., Kim, J., Yeom, M., Lee, I. C., Nam, H. G. (2007). A GUS/Luciferase fusion reporter for plant gene trapping and for assay of promoter activity with luciferin-dependent control of the reporter protein stability. Plant Cell Physiol. 48, 1121–1131. doi: 10.1093/pcp/pcm081
Li, X. J., Cao, L. J., Jiao, B. B., Yang, H. F., Ma, C. S., Liang, Y. (2022). The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion (Allium cepa l.). Hortic. RES-ENGLAND 9, 128. doi: 10.1093/hr/uhac128
Li, G. W., Chen, D. Y., Tang, X. F., Liu, Y. S. (2018). Heterologous expression of kiwifruit (Actinidia chinensis) GOLDEN2-LIKE homolog elevates chloroplast level and nutritional quality in tomato (Solanum lycopersicum). PLANTA 247, 1351–1362. doi: 10.1007/s00425-018-2853-6
Li, Y., Li, W. J., Wang, Y. K., Liang, R. F., Don, C. Z. (2007). Study on inheritance of fruit colour in chieh-qua. Northern Hortic. 2007 (10), 14–15. doi: 10.13989/j.cnki.0517-6611.2008.31.106
Li, X. J., Qian, W. Q., Zhao, Y. S., Wang, C. L., Shen, J., Zhu, J. K., et al. (2012). Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. P Natl. Acad. Sci. U.S.A. 109, 11425–11430. doi: 10.1073/pnas.120855710
Li, M. F., Zhang, H., Yang, Y., Wang, H., Xue, Z., Fan, Y. W., et al. (2022). Rosa1, a transposable element-like insertion, produces red petal coloration in rose through altering RcMYB114 transcription. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.857684
Li, B. B., Zhao, S. J., Dou, J. L., Ali, A., Gebremeskel, H., Gao, L., et al. (2019). Genetic mapping and development of molecular markers for a candidate gene locus controlling rind color in watermelon. Theor. Appl. Genet. 132, 2741–2753. doi: 10.1007/s00122-019-03384-3
Liu, H. Q., Jiao, J. Q., Liang, X. J., Liu, J., Meng, H. W., Chen, S. X., et al. (2016). Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus l.). Theor. Appl. Genet. 129, 1247–1256. doi: 10.1007/s00122-016-2700-8
Liu, H. Q., Meng, H. W., Pan, Y. P., Liang, X. J., Jiao, J. Q., Li, Y. H., et al. (2015). Fine genetic mapping of the white immature fruit color gene w to a 33.0-kb region in cucumber (Cucumis sativus l.). Theor. Appl. Genet. 128, 2375–2385. doi: 10.1007/s00122-015-2592-z
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. METHODS 25, 402–408. doi: 10.1006/meth.2001.1262
Lun, Y. Y., Wang, X., Zhang, C. Z., Yang, L., Gao, D. L., Chen, H. M., et al. (2016). A CsYcf54 variant conferring light green coloration in cucumber. EUPHYTICA 208, 509–517. doi: 10.1007/s10681-015-1592-z
Ma, L. L., Liu, Z. G., Cheng, Z. K., Gou, J. Q., Chen, J. Y., Yu, W. J., et al. (2021). Identification and application of BhAPRR2 controlling peel colour in wax gourd (Benincasa hispida). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.716772
Meggers, B. J., Ayensu, E. S., Duckworth, W. D., Biology, A. F. T. (1973). Tropical forest ecosystems in Africa and south america; a comparative review (Washington: Smithsonian Institution Press).
Nadakuduti, S. S., Holdsworth, W. L., Klein, C. L., Barry, C. S. (2014). KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 78, 1022–1033. doi: 10.1111/tpj.12529
Nadeem, M. A., Nawaz, M. A., Shahid, M. Q., Dogan, Y., Comertpay, G., Yildiz, M., et al. (2018). DNA Molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. BIOTEC EQ 32, 261–285. doi: 10.1080/13102818.2017.1400401
Nguyen, C. V., Vrebalov, J. T., Gapper, N. E., Zheng, Y., Zhong, S. L., Fei, Z. J., et al. (2014). Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26, 585–601. doi: 10.1105/tpc.113.118794
Ni, F. R., Wu, L., Wang, Q., Hong, J., Qi, Y. J., Zhou, X. P. (2017). Turnip yellow mosaic virus P69 interacts with and suppresses GLK transcription factors to cause pale-green symptoms in Arabidopsis. Mol. Plant 10, 764–766. doi: 10.1016/j.molp.2016.12.003
Ou, D. D. (2019). Genetic analysis and gene mapping for green peel color in melon (Cucumis melo l.) (China: Chinese Academy of Agricultural Sciences).
Pan, Y., Bradley, G., Pyke, K., Ball, G., Lu, C. G., Fray, R., et al. (2013). Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 161, 1476–1485. doi: 10.1104/pp.112.212654
Powell, A., Nguyen, C. V., Hill, T., Cheng, K. L., Figueroa-Balderas, R., Aktas, H., et al. (2012). Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. SCIENCE 336, 1711–1715. doi: 10.1126/science.1222218
Qing, C., HaoRu, T., XiaoLi, D., YanXia, H., Ya, L., Yan, J., et al. (2009). Progress in the study of plant myb transcription factors. Genomics Appl. Biol. 28, 365–372. doi: 10.3969/gab.028.000365
Rossini, L., Cribb, L., Martin, D. J., Langdale, J. A. (2001). The maize Golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 13, 1231–1244. doi: 10.1105/tpc.13.5.1231
Takos, A. M., Jaffe, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Wang, M., Chen, L., Liang, Z. J., He, X. M., Liu, W. R., Jiang, B., et al. (2020). Metabolome and transcriptome analyses reveal chlorophyll and anthocyanin metabolism pathway associated with cucumber fruit skin color. BMC Plant Biol. 20. doi: 10.1186/s12870-020-02597-9
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., Langdale, J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128. doi: 10.1105/tpc.108.065250
Whitaker, T. W. (1971). “Endemism and pre-Columbian migration of bottle gourd, lagenaria siceraria. mol, standl,” in Man across the sea. Ed. Riley, C. L. (Austin: University of Texas Press), 78–218. doi: 10.7560/701175-019
Wilkie, S., Clark, M. S., Leroy, P., Merlino, M., Nègre, S., Caissard, J. C., et al. (1997). “Genomic DNA isolation, southern blotting and hybridization,” in Plant molecular biology — a laboratory manual Springer, Berlin, Heidelberg: Springer, 3–53. doi: 10.1007/978-3-642-87873-2_1
Xu, D. D., Sun, D., Diao, Y. L., Liu, M. X., Gao, J., Wu, B., et al. (2019). Fast mapping of a chlorophyll b synthesis-deficiency gene in barley (Hordeum vulgare l.) via bulked-segregant analysis with reduced-representation sequencing. Crop J. 7, 58–64. doi: 10.1016/j.cj.2018.07.002
Xu, P., Wang, Y., Sun, F. S., Wu, R. L., Du, H. L., Wang, Y. H., et al. (2021). Long-read genome assembly and genetic architecture of fruit shape in the bottle gourd. Plant J. 107, 956–968. doi: 10.1111/tpj.15358
Zhao, G. W., Lian, Q., Zhang, Z. H., Fu, Q. S., He, Y. H., Ma, S., et al. (2019). A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 51, 1607. doi: 10.1038/s41588-019-0522-8
Zhou, Q., Wang, S. H., Hu, B. W., Chen, H. M., Zhang, Z. H., Huang, S. W. (2015). An ACCUMULATION AND REPLICATION OF CHLOROPLASTS 5 gene mutation confers light green peel in cucumber. J. Integr. Plant Biol. 57, 936–942. doi: 10.1111/jipb.12355
Zhu, X. Y., Chen, J. Y., Xie, Z. K., Gao, J., Ren, G. D., Gao, S., et al. (2015). Jasmonic acid promotes degreening via MYC2/3/4-and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84, 597–610. doi: 10.1111/tpj.13030
Zhu, D. D., Liu, Z. G., Bao, Z. Y., Jiao, X. X., Wang, X. Y. (2016). Analysis of ISSR markers linked to the fruit color trait gene in chieh-qua. Genomics Appl. Biol. 35, 2781–2787. doi: 10.13417/j.gab.035.002781
Keywords: bottle gourd, rind color, chlorophyll, APRR2, promoter variation, race
Citation: Huo Y, Zhang G, Yu W, Liu Z, Shen M, Zhao R, Hu S, Zheng X, Wang P and Yang Y (2023) Forward genetic studies reveal LsAPRR2 as a key gene in regulating the green color of pericarp in bottle gourd (Lagenaria siceraria). Front. Plant Sci. 14:1130669. doi: 10.3389/fpls.2023.1130669
Received: 23 December 2022; Accepted: 31 January 2023;
Published: 15 February 2023.
Edited by:
Honghao Lv, Insititute of Vegetables and Flowers (CAAS), ChinaReviewed by:
Changlong Wen, Beijing Vegetable Research Center, ChinaKailiang Bo, Insititute of Vegetables and Flowers (CAAS), China
Copyright © 2023 Huo, Zhang, Yu, Liu, Shen, Zhao, Hu, Zheng, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjuan Yang, yjyang85@126.com; Peng Wang, wangpeng@gxaas.net
 Yulai Huo
Yulai Huo Gui Zhang
Gui Zhang Peng Wang
Peng Wang