Induced Pluripotent Stem Cell-Derived Organoids: Their Implication in COVID-19 Modeling
Abstract
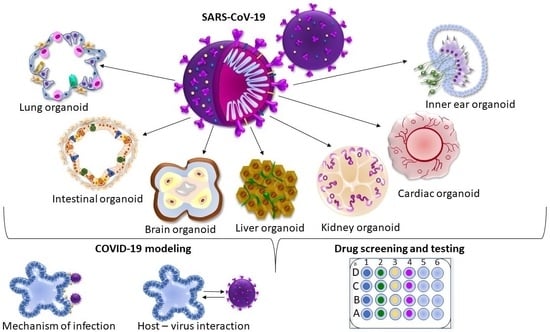
:1. Introduction
2. iPSC-Derived Organoids in COVID-19 Modeling
2.1. iPSC-Derived Airway Organoids
2.2. iPSC-Derived Brain Organoids
2.3. iPSC-Derived Intestinal Organoids
2.4. iPSC-Derived Liver Organoids
2.5. iPSC-Derived Cardiac and Blood Vessel Organoids
2.6. iPSC-Derived Kidney Organoids
2.7. iPSC-Derived Inner Ear Organoids
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/?mapFilter=deaths (accessed on 22 December 2022).
- Chakrabarty, K.; Shetty, R.; Argulwar, S.; Das, D.; Ghosh, A. Induced pluripotent stem cell-based disease modeling and prospective immune therapy for coronavirus disease 2019. Cytotherapy 2022, 24, 235–248. [Google Scholar]
- Marchiano, S.; Hsiang, T.Y.; Khanna, A.; Higashi, T.; Whitmore, L.S.; Bargehr, J.; Davaapil, H.; Chang, J.; Smith, E.; Ong, L.P.; et al. SARS-CoV-2 Infects Human Pluripotent Stem Cell-Derived Cardiomyocytes, Impairing Electrical and Mechanical Function. Stem Cell Rep. 2021, 16, 478–492. [Google Scholar] [CrossRef]
- Lei, H.Y.; Ding, Y.H.; Nie, K.; Dong, Y.M.; Xu, J.H.; Yang, M.L.; Liu, M.Q.; Wei, L.; Nasser, M.I.; Xu, L.Y.; et al. Potential effects of SARS-CoV-2 on the gastrointestinal tract and liver. Biomed. Pharmacother. 2021, 133, 111064. [Google Scholar]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L.; et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020, 69, 997–1001. [Google Scholar] [PubMed]
- Abboud, H.; Abboud, F.Z.; Kharbouch, H.; Arkha, Y.; El Abbadi, N.; El Ouahabi, A. COVID-19 and SARS-Cov-2 Infection: Pathophysiology and Clinical Effects on the Nervous System. World Neurosurg. 2020, 140, 49–53. [Google Scholar] [CrossRef]
- Brola, W.; Wilski, M. Neurological consequences of COVID-19. Pharmacol. Rep. 2022, 74, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef]
- Li, M.; Wang, H.; Tian, L.; Pang, Z.; Yang, Q.; Huang, T.; Fan, J.; Song, L.; Tong, Y.; Fan, H. COVID-19 vaccine development: Milestones, lessons and prospects. Signal Transduct. Target. Ther. 2022, 7, 146. [Google Scholar]
- Motamedi, H.; Ari, M.M.; Dashtbin, S.; Fathollahi, M.; Hossainpour, H.; Alvandi, A.; Moradi, J.; Abiri, R. An update review of globally reported SARS-CoV-2 vaccines in preclinical and clinical stages. Int. Immunopharmacol. 2021, 96, 107763. [Google Scholar]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248. [Google Scholar]
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021, 6, 1219–1232. [Google Scholar] [CrossRef]
- Nchioua, R.; Schundner, A.; Kmiec, D.; Prelli Bozzo, C.; Zech, F.; Koepke, L.; Graf, A.; Krebs, S.; Blum, H.; Frick, M.; et al. SARS-CoV-2 Variants of Concern Hijack IFITM2 for Efficient Replication in Human Lung Cells. J. Virol. 2022, 96, e0059422. [Google Scholar]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef]
- Yang, L.; Gou, J.; Gao, J.; Huang, L.; Zhu, Z.; Ji, S.; Liu, H.; Xing, L.; Yao, M.; Zhang, Y. Immune characteristics of severe and critical COVID-19 patients. Signal Transduct. Target. Ther. 2020, 5, 179. [Google Scholar] [CrossRef]
- Conti, P.; Ronconi, G.; Caraffa, A.; Gallenga, C.E.; Ross, R.; Frydas, I.; Kritas, S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents 2020, 34, 327–331. [Google Scholar]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar]
- Aid, M.; Busman-Sahay, K.; Vidal, S.J.; Maliga, Z.; Bondoc, S.; Starke, C.; Terry, M.; Jacobson, C.A.; Wrijil, L.; Ducat, S.; et al. Vascular Disease and Thrombosis in SARS-CoV-2-Infected Rhesus Macaques. Cell 2020, 183, 1354–1366.e1313. [Google Scholar]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar]
- Chan, J.F.; Zhang, A.J.; Yuan, S.; Poon, V.K.; Chan, C.C.; Lee, A.C.; Chan, W.M.; Fan, Z.; Tsoi, H.W.; Wen, L.; et al. Simulation of the Clinical and Pathological Manifestations of Coronavirus Disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for Disease Pathogenesis and Transmissibility. Clin. Infect. Dis. 2020, 71, 2428–2446. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Oude Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar]
- Winkler, E.S.; Chen, R.E.; Alam, F.; Yildiz, S.; Case, J.B.; Uccellini, M.B.; Holtzman, M.J.; Garcia-Sastre, A.; Schotsaert, M.; Diamond, M.S. SARS-CoV-2 Causes Lung Infection without Severe Disease in Human ACE2 Knock-In Mice. J. Virol. 2022, 96, e0151121. [Google Scholar] [CrossRef]
- de Melo, B.A.G.; Benincasa, J.C.; Cruz, E.M.; Maricato, J.T.; Porcionatto, M.A. 3D culture models to study SARS-CoV-2 infectivity and antiviral candidates: From spheroids to bioprinting. Biomed. J. 2021, 44, 31–42. [Google Scholar]
- Xia, Y.; Carpentier, A.; Cheng, X.; Block, P.D.; Zhao, Y.; Zhang, Z.; Protzer, U.; Liang, T.J. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J. Hepatol. 2017, 66, 494–503. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, Y.; Nan, Y.; Qiao, L. Induced pluripotent stem cells (iPSCs) in the modeling of hepatitis C virus infection. Curr. Stem. Cell Res. Ther. 2015, 10, 216–219. [Google Scholar] [CrossRef]
- Majolo, F.; Marinowic, D.R.; Moura, A.; Machado, D.C.; da Costa, J.C. Use of induced pluripotent stem cells (iPSCs) and cerebral organoids in modeling the congenital infection and neuropathogenesis induced by Zika virus. J. Med. Virol. 2019, 91, 525–532. [Google Scholar]
- D’Aiuto, L.; Bloom, D.C.; Naciri, J.N.; Smith, A.; Edwards, T.G.; McClain, L.; Callio, J.A.; Jessup, M.; Wood, J.; Chowdari, K.; et al. Modeling Herpes Simplex Virus 1 Infections in Human Central Nervous System Neuronal Cells Using Two- and Three-Dimensional Cultures Derived from Induced Pluripotent Stem Cells. J. Virol. 2019, 93, e00111-19. [Google Scholar] [CrossRef]
- Finkbeiner, S.R.; Zeng, X.L.; Utama, B.; Atmar, R.L.; Shroyer, N.F.; Estes, M.K. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 2012, 3, e00159-12. [Google Scholar]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.E.; Wang, P.; Duan, F.; Tang, X.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [Google Scholar]
- Jansen, J.; Reimer, K.C.; Nagai, J.S.; Varghese, F.S.; Overheul, G.J.; de Beer, M.; Roverts, R.; Daviran, D.; Fermin, L.A.S.; Willemsen, B.; et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell 2022, 29, 217–231.e218. [Google Scholar]
- Bose, B.; Kapoor, S.; Nihad, M. Induced Pluripotent Stem Cell Derived Human Lung Organoids to Map and Treat the SARS-CoV2 Infections In Vitro. Adv. Exp. Med. Biol. 2021, 1312, 1–17. [Google Scholar]
- Aboul-Soud, M.A.M.; Alzahrani, A.J.; Mahmoud, A. Induced Pluripotent Stem Cells (iPSCs)-Roles in Regenerative Therapies, Disease Modelling and Drug Screening. Cells 2021, 10, 2319. [Google Scholar] [CrossRef]
- de Dios-Figueroa, G.T.; Aguilera-Marquez, J.D.R.; Camacho-Villegas, T.A.; Lugo-Fabres, P.H. 3D Cell Culture Models in COVID-19 Times: A Review of 3D Technologies to Understand and Accelerate Therapeutic Drug Discovery. Biomedicines 2021, 9, 602. [Google Scholar] [CrossRef]
- Pei, R.; Feng, J.; Zhang, Y.; Sun, H.; Li, L.; Yang, X.; He, J.; Xiao, S.; Xiong, J.; Lin, Y.; et al. Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection. Protein Cell 2021, 12, 717–733. [Google Scholar] [CrossRef]
- Samuel, R.M.; Majd, H.; Richter, M.N.; Ghazizadeh, Z.; Zekavat, S.M.; Navickas, A.; Ramirez, J.T.; Asgharian, H.; Simoneau, C.R.; Bonser, L.R.; et al. Androgen Signaling Regulates SARS-CoV-2 Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. Cell Stem Cell 2020, 27, 876–889.e812. [Google Scholar] [CrossRef]
- Leeman, K.T.; Pessina, P.; Lee, J.H.; Kim, C.F. Mesenchymal Stem Cells Increase Alveolar Differentiation in Lung Progenitor Organoid Cultures. Sci. Rep. 2019, 9, 6479. [Google Scholar]
- Xu, G.; Li, Y.; Zhang, S.; Peng, H.; Wang, Y.; Li, D.; Jin, T.; He, Z.; Tong, Y.; Qi, C.; et al. SARS-CoV-2 promotes RIPK1 activation to facilitate viral propagation. Cell Res. 2021, 31, 1230–1243. [Google Scholar] [CrossRef]
- Salahudeen, A.A.; Choi, S.S.; Rustagi, A.; Zhu, J.; van Unen, V.; de la, O.S.; Flynn, R.A.; Margalef-Català, M.; Santos, A.J.M.; Ju, J.; et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 2020, 588, 670–675. [Google Scholar]
- Sato, T.; Morita, M.; Tanaka, R.; Inoue, Y.; Nomura, M.; Sakamoto, Y.; Miura, K.; Ito, S.; Sato, I.; Tanaka, N.; et al. Ex vivo model of non-small cell lung cancer using mouse lung epithelial cells. Oncol. Lett. 2017, 14, 6863–6868. [Google Scholar] [PubMed] [Green Version]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.; Tsai, Y.H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, 4, e05098. [Google Scholar] [PubMed]
- Wilkinson, D.C.; Alva-Ornelas, J.A.; Sucre, J.M.; Vijayaraj, P.; Durra, A.; Richardson, W.; Jonas, S.J.; Paul, M.K.; Karumbayaram, S.; Dunn, B.; et al. Development of a Three-Dimensional Bioengineering Technology to Generate Lung Tissue for Personalized Disease Modeling. Stem. Cells Transl. Med. 2017, 6, 622–633. [Google Scholar]
- Chen, Y.W.; Huang, S.X.; de Carvalho, A.; Ho, S.H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.L.; Bhattacharya, J.; et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef]
- Jose, S.S.; De Zuani, M.; Tidu, F.; Hortová Kohoutková, M.; Pazzagli, L.; Forte, G.; Spaccapelo, R.; Zelante, T.; Frič, J. Comparison of two human organoid models of lung and intestinal inflammation reveals Toll-like receptor signalling activation and monocyte recruitment. Clin. Transl. Immunol. 2020, 9, e1131. [Google Scholar] [CrossRef]
- Hein, R.F.C.; Conchola, A.S.; Fine, A.S.; Xiao, Z.; Frum, T.; Brastrom, L.K.; Akinwale, M.A.; Childs, C.J.; Tsai, Y.H.; Holloway, E.M.; et al. Stable iPSC-derived NKX2-1+ lung bud tip progenitor organoids give rise to airway and alveolar cell types. Development 2022, 149, dev200693. [Google Scholar]
- Leibel, S.L.; McVicar, R.N.; Winquist, A.M.; Snyder, E.Y. Generation of 3D Whole Lung Organoids from Induced Pluripotent Stem Cells for Modeling Lung Developmental Biology and Disease. J. Vis. Exp. 2021, 170, e62456. [Google Scholar] [CrossRef]
- Oglesby, I.K.; Schweikert, A.; Fox, B.; Redmond, C.; Donnelly, S.C.; Hurley, K. Lung organoids and other preclinical models of pulmonary fibrosis. Qjm 2021, 114, 167–173. [Google Scholar] [CrossRef]
- Suezawa, T.; Kanagaki, S.; Moriguchi, K.; Masui, A.; Nakao, K.; Toyomoto, M.; Tamai, K.; Mikawa, R.; Hirai, T.; Murakami, K.; et al. Disease modeling of pulmonary fibrosis using human pluripotent stem cell-derived alveolar organoids. Stem Cell Rep. 2021, 16, 2973–2987. [Google Scholar]
- Kim, J.H.; An, G.H.; Kim, J.Y.; Rasaei, R.; Kim, W.J.; Jin, X.; Woo, D.H.; Han, C.; Yang, S.R.; Kim, J.H.; et al. Human pluripotent stem-cell-derived alveolar organoids for modeling pulmonary fibrosis and drug testing. Cell Death Discov. 2021, 7, 48. [Google Scholar] [CrossRef]
- Korogi, Y.; Gotoh, S.; Ikeo, S.; Yamamoto, Y.; Sone, N.; Tamai, K.; Konishi, S.; Nagasaki, T.; Matsumoto, H.; Ito, I.; et al. In Vitro Disease Modeling of Hermansky-Pudlak Syndrome Type 2 Using Human Induced Pluripotent Stem Cell-Derived Alveolar Organoids. Stem Cell Rep. 2019, 13, 235. [Google Scholar] [CrossRef]
- Firth, A.L.; Menon, T.; Parker, G.S.; Qualls, S.J.; Lewis, B.M.; Ke, E.; Dargitz, C.T.; Wright, R.; Khanna, A.; Gage, F.H.; et al. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep. 2015, 12, 1385–1390. [Google Scholar]
- Kunisaki, S.M.; Jiang, G.; Biancotti, J.C.; Ho, K.K.Y.; Dye, B.R.; Liu, A.P.; Spence, J.R. Human induced pluripotent stem cell-derived lung organoids in an ex vivo model of the congenital diaphragmatic hernia fetal lung. Stem Cells Transl. Med. 2021, 10, 98–114. [Google Scholar]
- Leibel, S.L.; Winquist, A.; Tseu, I.; Wang, J.; Luo, D.; Shojaie, S.; Nathan, N.; Snyder, E.; Post, M. Reversal of Surfactant Protein B Deficiency in Patient Specific Human Induced Pluripotent Stem Cell Derived Lung Organoids by Gene Therapy. Sci. Rep. 2019, 9, 13450. [Google Scholar]
- Jacob, A.; Morley, M.; Hawkins, F.; McCauley, K.B.; Jean, J.C.; Heins, H.; Na, C.L.; Weaver, T.E.; Vedaie, M.; Hurley, K.; et al. Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell Stem Cell 2017, 21, 472–488.e410. [Google Scholar]
- Tiwari, S.K.; Wang, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Revealing Tissue-Specific SARS-CoV-2 Infection and Host Responses using Human Stem Cell-Derived Lung and Cerebral Organoids. Stem Cell Rep. 2021, 16, 437–445. [Google Scholar] [CrossRef]
- Spitalieri, P.; Centofanti, F.; Murdocca, M.; Scioli, M.G.; Latini, A.; Di Cesare, S.; Citro, G.; Rossi, A.; Orlandi, A.; Miersch, S.; et al. Two Different Therapeutic Approaches for SARS-CoV-2 in hiPSCs-Derived Lung Organoids. Cells 2022, 11, 1235. [Google Scholar]
- Djidrovski, I.; Georgiou, M.; Hughes, G.L.; Patterson, E.I.; Casas-Sanchez, A.; Pennington, S.H.; Biagini, G.A.; Moya-Molina, M.; van den Bor, J.; Smit, M.J.; et al. SARS-CoV-2 infects an upper airway model derived from induced pluripotent stem cells. Stem Cells 2021, 39, 1310–1321. [Google Scholar]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020, 30, 343–355. [Google Scholar]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Han, Y.; Yang, L.; Lacko, L.A.; Chen, S. Human organoid models to study SARS-CoV-2 infection. Nat. Methods 2022, 19, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Sano, E.; Suzuki, T.; Hashimoto, R.; Itoh, Y.; Sakamoto, A.; Sakai, Y.; Saito, A.; Okuzaki, D.; Motooka, D.; Muramoto, Y.; et al. Cell response analysis in SARS-CoV-2 infected bronchial organoids. Commun. Biol. 2022, 5, 516. [Google Scholar]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [PubMed]
- Kadoshima, T.; Sakaguchi, H.; Nakano, T.; Soen, M.; Ando, S.; Eiraku, M.; Sasai, Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 2013, 110, 20284–20289. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef] [PubMed]
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015, 10, 537–550. [Google Scholar] [CrossRef]
- Garcez, P.P.; Loiola, E.C.; Madeiro da Costa, R.; Higa, L.M.; Trindade, P.; Delvecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [PubMed]
- Chen, X.; Sun, G.; Tian, E.; Zhang, M.; Davtyan, H.; Beach, T.G.; Reiman, E.M.; Blurton-Jones, M.; Holtzman, D.M.; Shi, Y. Modeling Sporadic Alzheimer’s Disease in Human Brain Organoids under Serum Exposure. Adv. Sci. 2021, 8, e2101462. [Google Scholar] [CrossRef]
- Logan, S.; Arzua, T.; Canfield, S.G.; Seminary, E.R.; Sison, S.L.; Ebert, A.D.; Bai, X. Studying Human Neurological Disorders Using Induced Pluripotent Stem Cells: From 2D Monolayer to 3D Organoid and Blood Brain Barrier Models. Compr. Physiol. 2019, 9, 565–611. [Google Scholar]
- Gomes, A.R.; Fernandes, T.G.; Cabral, J.M.S.; Diogo, M.M. Modeling Rett Syndrome with Human Pluripotent Stem Cells: Mechanistic Outcomes and Future Clinical Perspectives. Int. J. Mol. Sci. 2021, 22, 3751. [Google Scholar] [PubMed]
- Tang, X.Y.; Xu, L.; Wang, J.; Hong, Y.; Wang, Y.; Zhu, Q.; Wang, D.; Zhang, X.Y.; Liu, C.Y.; Fang, K.H.; et al. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J. Clin. Investig. 2021, 131, e135763. [Google Scholar] [PubMed]
- Ao, Z.; Cai, H.; Wu, Z.; Song, S.; Karahan, H.; Kim, B.; Lu, H.C.; Kim, J.; Mackie, K.; Guo, F. Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab. Chip. 2021, 21, 2751–2762. [Google Scholar] [PubMed]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e939. [Google Scholar] [PubMed]
- Bullen, C.K.; Hogberg, H.T.; Bahadirli-Talbott, A.; Bishai, W.R.; Hartung, T.; Keuthan, C.; Looney, M.M.; Pekosz, A.; Romero, J.C.; Sillé, F.C.M.; et al. Infectability of human BrainSphere neurons suggests neurotropism of SARS-CoV-2. Altex 2020, 37, 665–671. [Google Scholar]
- Pellegrini, L.; Albecka, A.; Mallery, D.L.; Kellner, M.J.; Paul, D.; Carter, A.P.; James, L.C.; Lancaster, M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 2020, 27, 951–961.e955. [Google Scholar]
- Mesci, P.; de Souza, J.S.; Martin-Sancho, L.; Macia, A.; Saleh, A.; Yin, X.; Snethlage, C.; Adams, J.W.; Avansini, S.H.; Herai, R.H.; et al. SARS-CoV-2 infects human brain organoids causing cell death and loss of synapses that can be rescued by treatment with Sofosbuvir. PLoS Biol. 2022, 20, e3001845. [Google Scholar] [CrossRef]
- Chen, D.; Su, X.; Chen, H.; Chen, S.; Zhao, Y.; Wei, W. Human Organoids as a Promising Platform for Fighting COVID-19. Int. J. Biol. Sci. 2022, 18, 901–910. [Google Scholar] [CrossRef]
- Ramani, A.; Müller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Müller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020, 39, e106230. [Google Scholar]
- Song, E.; Zhang, C.; Israelow, B.; Lu-Culligan, A.; Prado, A.V.; Skriabine, S.; Lu, P.; Weizman, O.E.; Liu, F.; Dai, Y.; et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, B.Z.; Chu, H.; Han, S.; Shuai, H.; Deng, J.; Hu, Y.F.; Gong, H.R.; Lee, A.C.; Zou, Z.; Yau, T.; et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020, 30, 928–931. [Google Scholar] [PubMed]
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, H.B.; Lyu, J.R.; Lei, X.M.; Li, W.; Wu, G.; Lyu, J.; Dai, Z.M. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect Dis. 2020, 96, 19–24. [Google Scholar]
- Mithal, A.; Hume, A.J.; Lindstrom-Vautrin, J.; Villacorta-Martin, C.; Olejnik, J.; Bullitt, E.; Hinds, A.; Mühlberger, E.; Mostoslavsky, G. Human Pluripotent Stem Cell-Derived Intestinal Organoids Model SARS-CoV-2 Infection Revealing a Common Epithelial Inflammatory Response. Stem Cell Rep. 2021, 16, 940–953. [Google Scholar]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar]
- Múnera, J.O.; Sundaram, N.; Rankin, S.A.; Hill, D.; Watson, C.; Mahe, M.; Vallance, J.E.; Shroyer, N.F.; Sinagoga, K.L.; Zarzoso-Lacoste, A.; et al. Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling. Cell Stem Cell 2017, 21, 51–64.e56. [Google Scholar] [CrossRef]
- Loffet, E.; Brossard, L.; Mahe, M.M. Pluripotent stem cell derived intestinal organoids with an enteric nervous system. Methods Cell Biol. 2020, 159, 175–199. [Google Scholar]
- Workman, M.J.; Mahe, M.M.; Trisno, S.; Poling, H.M.; Watson, C.L.; Sundaram, N.; Chang, C.F.; Schiesser, J.; Aubert, P.; Stanley, E.G.; et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017, 23, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.L.; Mahe, M.M.; Múnera, J.; Howell, J.C.; Sundaram, N.; Poling, H.M.; Schweitzer, J.I.; Vallance, J.E.; Mayhew, C.N.; Sun, Y.; et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014, 20, 1310–1314. [Google Scholar] [CrossRef]
- Crespo, M.; Vilar, E.; Tsai, S.Y.; Chang, K.; Amin, S.; Srinivasan, T.; Zhang, T.; Pipalia, N.H.; Chen, H.J.; Witherspoon, M.; et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 2017, 23, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Onozato, D.; Akagawa, T.; Kida, Y.; Ogawa, I.; Hashita, T.; Iwao, T.; Matsunaga, T. Application of Human Induced Pluripotent Stem Cell-Derived Intestinal Organoids as a Model of Epithelial Damage and Fibrosis in Inflammatory Bowel Disease. Biol. Pharm. Bull. 2020, 43, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Kandilogiannakis, L.; Filidou, E.; Drygiannakis, I.; Tarapatzi, G.; Didaskalou, S.; Koffa, M.; Arvanitidis, K.; Bamias, G.; Valatas, V.; Paspaliaris, V.; et al. Development of a Human Intestinal Organoid Model for In Vitro Studies on Gut Inflammation and Fibrosis. Stem Cells Int. 2021, 2021, 9929461. [Google Scholar] [CrossRef]
- Gleeson, J.P.; Estrada, H.Q.; Yamashita, M.; Svendsen, C.N.; Targan, S.R.; Barrett, R.J. Development of Physiologically Responsive Human iPSC-Derived Intestinal Epithelium to Study Barrier Dysfunction in IBD. Int. J. Mol. Sci. 2020, 21, 1438. [Google Scholar]
- Estrada, H.Q.; Patel, S.; Rabizadeh, S.; Casero, D.; Targan, S.R.; Barrett, R.J. Development of a Personalized Intestinal Fibrosis Model Using Human Intestinal Organoids Derived From Induced Pluripotent Stem Cells. Inflamm. Bowel. Dis. 2022, 28, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Jurickova, I.; Bonkowski, E.; Angerman, E.; Novak, E.; Huron, A.; Akers, G.; Iwasawa, K.; Braun, T.; Hadar, R.; Hooker, M.; et al. Eicosatetraynoic Acid and Butyrate Regulate Human Intestinal Organoid Mitochondrial and Extracellular Matrix Pathways Implicated in Crohn’s Disease Strictures. Inflamm. Bowel. Dis. 2022, 28, 988–1003. [Google Scholar] [CrossRef] [PubMed]
- Lui, K.N.; Ngan, E.S. Human Pluripotent Stem Cell-Based Models for Hirschsprung Disease: From 2-D Cell to 3-D Organoid Model. Cells 2022, 11, 3428. [Google Scholar] [CrossRef]
- Sarvestani, S.K.; Signs, S.; Hu, B.; Yeu, Y.; Feng, H.; Ni, Y.; Hill, D.R.; Fisher, R.C.; Ferrandon, S.; DeHaan, R.K.; et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat. Commun. 2021, 12, 262. [Google Scholar]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Krüger, J.; Groß, R.; Conzelmann, C.; Müller, J.A.; Koepke, L.; Sparrer, K.M.J.; Weil, T.; Schütz, D.; Seufferlein, T.; Barth, T.F.E.; et al. Drug Inhibition of SARS-CoV-2 Replication in Human Pluripotent Stem Cell-Derived Intestinal Organoids. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 935–948. [Google Scholar] [PubMed]
- Zhao, X.; Li, C.; Liu, X.; Chiu, M.C.; Wang, D.; Wei, Y.; Chu, H.; Cai, J.P.; Hau-Yee Chan, I.; Kak-Yuen Wong, K.; et al. Human Intestinal Organoids Recapitulate Enteric Infections of Enterovirus and Coronavirus. Stem Cell Rep. 2021, 16, 493–504. [Google Scholar]
- Wang, Y.; Liu, S.; Liu, H.; Li, W.; Lin, F.; Jiang, L.; Li, X.; Xu, P.; Zhang, L.; Zhao, L.; et al. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J. Hepatol. 2020, 73, 807–816. [Google Scholar]
- Richards, A.; Friesen, M.; Khalil, A.; Barrasa, M.I.; Gehrke, L.; Jaenisch, R. SARS-CoV-2 infection of human pluripotent stem cell-derived liver organoids reveals potential mechanisms of liver pathology. iScience 2022, 25, 105146. [Google Scholar] [PubMed]
- Yang, L.; Han, Y.; Nilsson-Payant, B.E.; Gupta, V.; Wang, P.; Duan, X.; Tang, X.; Zhu, J.; Zhao, Z.; Jaffré, F.; et al. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 2020, 27, 125–136.e127. [Google Scholar] [PubMed]
- Olgasi, C.; Cucci, A.; Follenzi, A. iPSC-Derived Liver Organoids: A Journey from Drug Screening, to Disease Modeling, Arriving to Regenerative Medicine. Int. J. Mol. Sci. 2020, 21, 6215. [Google Scholar] [CrossRef]
- Gordillo, M.; Evans, T.; Gouon-Evans, V. Orchestrating liver development. Development 2015, 142, 2094–2108. [Google Scholar] [CrossRef]
- Takebe, T.; Zhang, R.R.; Koike, H.; Kimura, M.; Yoshizawa, E.; Enomura, M.; Koike, N.; Sekine, K.; Taniguchi, H. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat. Protoc. 2014, 9, 396–409. [Google Scholar]
- Wu, F.; Wu, D.; Ren, Y.; Huang, Y.; Feng, B.; Zhao, N.; Zhang, T.; Chen, X.; Chen, S.; Xu, A. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 2019, 70, 1145–1158. [Google Scholar]
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Shrike Zhang, Y.; Shin, S.R.; Calzone, G.; et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 014101. [Google Scholar] [CrossRef]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered Liver-on-a-Chip Platform to Mimic Liver Functions and Its Biomedical Applications: A Review. Micromachines 2019, 10, 676. [Google Scholar] [PubMed]
- Ma, L.D.; Wang, Y.T.; Wang, J.R.; Wu, J.L.; Meng, X.S.; Hu, P.; Mu, X.; Liang, Q.L.; Luo, G.A. Design and fabrication of a liver-on-a-chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab. Chip. 2018, 18, 2547–2562. [Google Scholar] [CrossRef] [PubMed]
- Ramli, M.N.B.; Lim, Y.S.; Koe, C.T.; Demircioglu, D.; Tng, W.; Gonzales, K.A.U.; Tan, C.P.; Szczerbinska, I.; Liang, H.; Soe, E.L.; et al. Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 2020, 159, 1471–1486.e1412. [Google Scholar]
- Shinozawa, T.; Kimura, M.; Cai, Y.; Saiki, N.; Yoneyama, Y.; Ouchi, R.; Koike, H.; Maezawa, M.; Zhang, R.R.; Dunn, A.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell-Derived Organoids. Gastroenterology 2021, 160, 831–846.e810. [Google Scholar] [CrossRef]
- Cao, D.; Ge, J.Y.; Wang, Y.; Oda, T.; Zheng, Y.W. Hepatitis B virus infection modeling using multicellular organoids derived from human induced pluripotent stem cells. World J. Gastroenterol. 2021, 27, 4784–4801. [Google Scholar] [PubMed]
- Zhao, B.; Ni, C.; Gao, R.; Wang, Y.; Yang, L.; Wei, J.; Lv, T.; Liang, J.; Zhang, Q.; Xu, W.; et al. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 2020, 11, 771–775. [Google Scholar] [CrossRef]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar]
- Lee, S.J.; Kim, H.A.; Kim, S.J.; Lee, H.A. Improving Generation of Cardiac Organoids from Human Pluripotent Stem Cells Using the Aurora Kinase Inhibitor ZM447439. Biomedicines 2021, 9, 1952. [Google Scholar] [CrossRef]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef]
- Richards, D.J.; Li, Y.; Kerr, C.M.; Yao, J.; Beeson, G.C.; Coyle, R.C.; Chen, X.; Jia, J.; Damon, B.; Wilson, R.; et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 2020, 4, 446–462. [Google Scholar] [CrossRef]
- Dias, T.P.; Pinto, S.N.; Carvalho, S.; Fernandes, T.G.; Fernandes, F.; Diogo, M.M.; Peleteiro, M.C.; Prieto, M.; Cabral, J.M.S. Cost-Effective Mechanical Aggregation of Cardiac Progenitors and Encapsulation in Matrigel Support Self-Organization in a Dynamic Culture Environment. Int. J. Mol. Sci. 2022, 23, 15785. [Google Scholar] [CrossRef] [PubMed]
- Yiangou, L.; Davis, R.P.; Mummery, C.L. Using Cardiovascular Cells from Human Pluripotent Stem Cells for COVID-19 Research: Why the Heart Fails. Stem Cell Rep. 2021, 16, 385–397. [Google Scholar]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e907. [Google Scholar] [CrossRef]
- Pérez-Bermejo, J.A.; Kang, S.; Rockwood, S.J.; Simoneau, C.R.; Joy, D.A.; Ramadoss, G.N.; Silva, A.C.; Flanigan, W.R.; Li, H.; Nakamura, K.; et al. SARS-CoV-2 infection of human iPSC-derived cardiac cells predicts novel cytopathic features in hearts of COVID-19 patients. bioRxiv 2020. [Google Scholar] [CrossRef]
- Choi, S.W.; Shin, J.S.; Park, S.J.; Jung, E.; Park, Y.G.; Lee, J.; Kim, S.J.; Park, H.J.; Lee, J.H.; Park, S.M.; et al. Antiviral activity and safety of remdesivir against SARS-CoV-2 infection in human pluripotent stem cell-derived cardiomyocytes. Antiviral. Res. 2020, 184, 104955. [Google Scholar] [CrossRef]
- Arhontoulis, D.C.; Kerr, C.; Richards, D.; Tjen, K.; Hyams, N.; Jones, J.A.; Deleon-Pennell, K.; Menick, D.; Lindner, D.; Westermann, D.; et al. Human Cardiac Organoids to Model COVID-19 Cytokine Storm Induced Cardiac Injuries. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chan, L.; Chaudhary, K.; Saha, A.; Chauhan, K.; Vaid, A.; Zhao, S.; Paranjpe, I.; Somani, S.; Richter, F.; Miotto, R.; et al. AKI in Hospitalized Patients with COVID-19. J. Am. Soc. Nephrol. 2021, 32, 151–160. [Google Scholar]
- Fisher, M.; Neugarten, J.; Bellin, E.; Yunes, M.; Stahl, L.; Johns, T.S.; Abramowitz, M.K.; Levy, R.; Kumar, N.; Mokrzycki, M.H.; et al. AKI in Hospitalized Patients with and without COVID-19: A Comparison Study. J. Am. Soc. Nephrol. 2020, 31, 2145–2157. [Google Scholar]
- Głowacka, M.; Lipka, S.; Młynarska, E.; Franczyk, B.; Rysz, J. Acute Kidney Injury in COVID-19. Int. J. Mol. Sci. 2021, 22, 8081. [Google Scholar] [CrossRef]
- Xia, S.; Wu, M.; Chen, S.; Zhang, T.; Ye, L.; Liu, J.; Li, H. Long Term Culture of Human Kidney Proximal Tubule Epithelial Cells Maintains Lineage Functions and Serves as an Ex vivo Model for Coronavirus Associated Kidney Injury. Virol. Sin. 2020, 35, 311–320. [Google Scholar] [CrossRef]
- Reimer, K.C.; Jansen, J.; Overheul, G.J.; Miesen, P.; van Rij, R.P.; Triana, S.H.; Smeets, B.; Schneider, R.K.; Kramann, R. Using human iPSC-derived kidney organoids to decipher SARS-CoV-2 pathology on single cell level. STAR Protoc. 2022, 3, 101612. [Google Scholar] [CrossRef] [PubMed]
- Vanslambrouck, J.M.; Wilson, S.B.; Tan, K.S.; Groenewegen, E.; Rudraraju, R.; Neil, J.; Lawlor, K.T.; Mah, S.; Scurr, M.; Howden, S.E.; et al. Enhanced metanephric specification to functional proximal tubule enables toxicity screening and infectious disease modelling in kidney organoids. Nat. Commun. 2022, 13, 5943. [Google Scholar] [CrossRef] [PubMed]
- Garreta, E.; Prado, P.; Stanifer, M.L.; Monteil, V.; Marco, A.; Ullate-Agote, A.; Moya-Rull, D.; Vilas-Zornoza, A.; Tarantino, C.; Romero, J.P.; et al. A diabetic milieu increases ACE2 expression and cellular susceptibility to SARS-CoV-2 infections in human kidney organoids and patient cells. Cell Metab. 2022, 34, 857–873.e9. [Google Scholar] [CrossRef] [PubMed]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Maier, B.; Baillie, G.J.; Ferguson, C.; Parton, R.G.; Wolvetang, E.J.; Roost, M.S.; Chuva de Sousa Lopes, S.M.; et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 2015, 526, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Kalejaiye, T.D.; Bhattacharya, R.; Burt, M.A.; Travieso, T.; Okafor, A.E.; Mou, X.; Blasi, M.; Musah, S. SARS-CoV-2 Employ BSG/CD147 and ACE2 Receptors to Directly Infect Human Induced Pluripotent Stem Cell-Derived Kidney Podocytes. Front. Cell Dev. Biol. 2022, 10, 855340. [Google Scholar] [PubMed]
- Vaira, L.A.; Salzano, G.; Deiana, G.; De Riu, G. Anosmia and Ageusia: Common Findings in COVID-19 Patients. Laryngoscope 2020, 130, 1787. [Google Scholar] [CrossRef]
- Meng, X.; Wang, J.; Sun, J.; Zhu, K. COVID-19 and Sudden Sensorineural Hearing Loss: A Systematic Review. Front. Neurol. 2022, 13, 883749. [Google Scholar]
- Fancello, V.; Fancello, G.; Hatzopoulos, S.; Bianchini, C.; Stomeo, F.; Pelucchi, S.; Ciorba, A. Sensorineural Hearing Loss Post-COVID-19 Infection: An Update. Audiol. Res. 2022, 12, 32. [Google Scholar] [CrossRef]
- Jeong, M.; Ocwieja, K.E.; Han, D.; Wackym, P.A.; Zhang, Y.; Brown, A.; Moncada, C.; Vambutas, A.; Kanne, T.; Crain, R.; et al. Direct SARS-CoV-2 infection of the human inner ear may underlie COVID-19-associated audiovestibular dysfunction. Commun. Med 2021, 1, 44. [Google Scholar] [CrossRef]
- Koehler, K.R.; Nie, J.; Longworth-Mills, E.; Liu, X.P.; Lee, J.; Holt, J.R.; Hashino, E. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 2017, 35, 583–589. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, H.; Lee, M.K.; Choi, W.H.; Jang, Y.; Shin, J.S.; Park, J.Y.; Bae, D.H.; Hyun, S.I.; Kim, K.H.; et al. Generation of human tonsil epithelial organoids as an ex vivo model for SARS-CoV-2 infection. Biomaterials 2022, 283, 121460. [Google Scholar] [CrossRef] [PubMed]
- Parihar, A.; Pandita, V.; Khan, R. 3D printed human organoids: High throughput system for drug screening and testing in current COVID-19 pandemic. Biotechnol. Bioeng. 2022, 119, 2669–2688. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, P.; Qin, J. Human Organoids and Organs-on-Chips for Addressing COVID-19 Challenges. Adv. Sci. 2022, 9, e2105187. [Google Scholar] [CrossRef] [PubMed]







| iPSC-Derived Organoids | Small Molecules, Growth Factors, and Recombinant Proteins Used for Organoid Differentiation | Permissiveness to SARS-CoV-2 | Pathological Features | Drug Screening | Ref. |
|---|---|---|---|---|---|
| Lung organoids | Activin A, SB431542, Noggin, SAG, FGF4, CHIR99021—foregut endoderm + FGF7, FGF10, CHIR99021, EGF, ATRA, and VEGF/PIGF—lung organoids |
Yes | Upregulated expressions of immune-regulatory genes, inflammation-activated inflammasome pathway | EK1 and camostat-inhibition of viral infection | [57] |
| Lung organoids | CHIR99021, rhKGF, dexamethasone, 8-Br-cAMP, 3-Isobutyl-1-methylxanthine—alveospheres | Yes | Features of cellular damage—alveolar cell hyperplasia with hyaline deposition | 15033-7 and DPP4 peptide-inhibition of viral infection | [58] |
| Brain and choroid plexus organoids | L 2-mercaptoethanol, dorsomorphin, A83-01, CHIR99021, SB431542, insulin, LDN-193189, SHH, and FGF8 | Yes—in choroid plexus cells; rarely in neurons and astrocytes | Proinflammatory cytokine response, cell death, formation of syncytia, damage of choroid plexus organoid integrity | - | [76] |
| BrainSphere organotypic model | BDNF protein, GDNF protein, dorsomorphin, heparin, and cyclopamine | Yes—few neurons | - | - | [77] |
| Brain and choroid plexus organoids | Heparin, Y27632, L 2-mercaptoethanol, insulin, and vitamin A, BMP4, and CHIR99021 | Yes—in choroid plexus surface lining cells | Damage of choroid plexus organoid integrity | - | [78] |
| Brain organoids | SB431542, dorsomorphin, Y27632, bFGF, EGF, BDNF protein, GDNF protein, L-ascorbic acid, and dibutyryl-cAMP | Yes | Cell death of cortical neurons, reduced number of excitatory synapses | Sofosbuvir—reduction of intracellular viral RNA levels | [79] |
| Brain organoids | Heparin, Y27632, L 2-mercaptoethanol, insulin, and vitamin A | Yes—only neurons | Presence of aberrant Tau protein in SARS-CoV-2 + neurons | - | [81] |
| Brain organoids | Heparin, Y27632, L 2-mercaptoethanol, insulin, and vitamin A | Yes—only neurons | Cell death of infected and neighboring cells | - | [82] |
| 3D neurospheres | Y27632, dorsomorphin, SB431542, FGF2, BDNF protein, GDNF protein, β-mercaptoethanol, and L-ascorbic acid | Yes—neural progenitor cells | Cell death | - | [83] |
| Enteroids Colonoids |
Dorsomorphin, SB431542, CHIR99021, rhBMP4, and retinoic acid-definitive endoderm + Y27632—final organoid |
Yes—in enterocytes and colonocytes | Upregulation of apoptosis-related genes, cellular stress, expression of inflammatory markers | - | [86] |
| Liver organoids | Activin A, BMP4, FGF4, CHIR9902, retinoic acid, HGF, and OSM | Yes—more in hepatocytes-like cells, less in cholangiocytes-like cells | Rapid change of gene expression in inflammatory signaling pathways | - | [105] |
| Liver organoids | Activin A, BMP4, bFGF, HGF, and OSM | Yes | Strong induction of inflammatory cytokines and chemokines | - | [106] |
| Blood vessel organoids | Y-27632, CHIR99021, BMP4, VEGF-A, FGF-2, and SB43152 | Yes | - | Human recombinant soluble ACE2-inhibition of viral infection | [124] |
| Cardiac organoids | Self-assembly process | Yes | Signs of fibrosis and reduced functions of cardiomyocytes, prothrombotic features of the vasculature–impaired nitric oxide-mediated endothelial function | - | [127] |
| Kidney organoids | CHIR99021, FGF9, and heparin | Yes—mainly in proximal tubules, less in loops of Henle | Upregulation of KIM-1 in proximal tubules | - | [133] |
| Kidney organoids | CHIR99021, FGF9, hEGF, and heparin | Yes—in podocytes and proximal tubules | Cell injury and dedifferentiation of infected cells, post-infection fibrosis | MAT-POS-b3e365b9-1—reduction of intracellular viral RNA levels | [33] |
| Diabetic kidney organoids | CHIR99021, FGF9, heparin, and activin A | Yes | Downregulation of glycolysis-related processes and increased inflammation | - | [134] |
| Inner ear organoids | BMP4, FGF-2, CHIR99021, LDN-193189, SB431542, and Y-27632 | Yes—vestibular hair cell-like cells | dsRNA presented in vestibular hair cell-like cells | - | [140] |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
|
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Csöbönyeiová, M.; Klein, M.; Kuniaková, M.; Varga, I.; Danišovič, Ľ. Induced Pluripotent Stem Cell-Derived Organoids: Their Implication in COVID-19 Modeling. Int. J. Mol. Sci. 2023, 24, 3459. https://doi.org/10.3390/ijms24043459
Csöbönyeiová M, Klein M, Kuniaková M, Varga I, Danišovič Ľ. Induced Pluripotent Stem Cell-Derived Organoids: Their Implication in COVID-19 Modeling. International Journal of Molecular Sciences. 2023; 24(4):3459. https://doi.org/10.3390/ijms24043459
Chicago/Turabian StyleCsöbönyeiová, Mária, Martin Klein, Marcela Kuniaková, Ivan Varga, and Ľuboš Danišovič. 2023. "Induced Pluripotent Stem Cell-Derived Organoids: Their Implication in COVID-19 Modeling" International Journal of Molecular Sciences 24, no. 4: 3459. https://doi.org/10.3390/ijms24043459