Transcriptome Analysis Reveals Candidate Genes Related to Anthocyanin Biosynthesis in Different Carrot Genotypes and Tissues
Abstract
:1. Introduction
2. Results
2.1. Total Anthocyanin Accumulation during Four Growth Stages
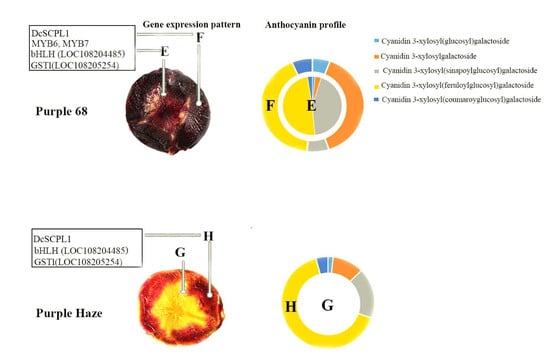
2.2. Characterization of Anthocyanins in the Carrot Phloem and Xylem
2.3. An Overview of Transcriptome Sequencing Datasets
2.4. Identification of Differentially Expressed Genes between the Phloem and Xylem of ‘Purple 68′ and ‘Purple Haze’
2.5. Differentially Expressed Structural Genes in Anthocyanin Biosynthesis Pathway
2.6. Candidate R2R3MYB and bHLH TFs Involved in Anthocyanin Biosynthesis
2.7. GST Involved in Anthocyanin Transport in Carrots
2.8. Validation of RNA-Seq Results with Quantitative Real-Time PCR (qRT-PCR)
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Total Anthocyanin Content and Individual Anthocyanin
4.3. RNA Extraction and cDNA Synthesis
4.4. Library Construction and Transcriptome Sequencing
4.5. RNA-Sequencing Data Analysis
4.6. RT-qPCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Zhai, R.; Wang, Z.; Zhang, S.; Meng, G.; Song, L.; Wang, Z.; Li, P.; Ma, F.; Xu, L. Two MYB transcription factors regulate flavonoid biosynthesis in pear fruit (Pyrus bretschneideri Rehd.) Rui. J. Exp. Bot. 2016, 67, 1275–1284. [Google Scholar] [CrossRef] [Green Version]
- Vavilov, N.; Dorofeev, V. Origin and Geography of Cultivated Plants. Available online: https://www.amazon.com/Origin-Geography-Cultivated-Plants-Vavilov/dp/0521111595 (accessed on 8 October 2019).
- Banga, O. Main Types of the Western Carotene Carrot and their Origin. Available online: https://www.amazon.com/types-western-carotene-carrot-origin/dp/B0006BY8VU (accessed on 8 October 2019).
- Simon, P.W. Domestication, Historical Development, and Modern Breeding of Carrot. Plant Breed. 2000, 19, 157–190. [Google Scholar]
- Vavilov, N. Centers of Origin of Cultivated Plants. pp. 495–496. Available online: https://www.researchgate.net/publication/313881301_On_Centers_of_Origin_of_Cultivated_Plants_Case_Study_of_Armenian_Highland (accessed on 8 October 2019).
- Giusti, M.; Wrolstad, R. Acylated anthocyanins from edible sources and their applications in food systems. Biochem. Eng. 2003, 14, 217–225. [Google Scholar] [CrossRef]
- Kammerer, D.; Carle, R.; Schieber, A. Characterization of phenolic acids in black carrots (Daucus carota ssp. sativus var. atrorubens Alef.) by high-performance liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Kurilich, A.C.; Clevidence, B.A.; Britz, S.J.; Simon, P.W.; Novotny, J.A. Plasma and urine responses are lower for acylated vs nonacylated anthocyanins from raw and cooked purple carrots. J. Agric. Food Chem. 2005, 53, 6537–6542. [Google Scholar] [CrossRef] [PubMed]
- Stintzing, F.C.; Carle, R. Functional properties of anthocyanins and betalains in plants, food, and in human nutrition. Trends Food Sci. Technol. 2004, 15, 19–38. [Google Scholar] [CrossRef]
- Montilla, E.C.; Arzaba, M.R.; Hillebrand, S.; Winterhalter, P. Anthocyanin Composition of Black Carrot (Daucus carota ssp. sativus var. atrorubens Alef.) Cultivars Antonina, Beta Sweet, Deep Purple, and ‘Purple Haze’. J. Agric. Food Chem. 2011, 59, 3385–3390. [Google Scholar] [CrossRef] [PubMed]
- Gras, C.C.; Bogner, H.; Carle, R.; Schweiggert, R.M. Effect of genuine non-anthocyanin phenolics and chlorogenic acid on color and stability of black carrot (Daucus carota ssp. sativus var. atrorubens Alef.) anthocyanins. Food Res. Int. 2016, 85, 291–300. [Google Scholar] [CrossRef]
- Kamiloglu, S.; Capanoglu, E.; Bilen, F.D.; Gonzales, G.B.; Grootaert, C.; Van De Wiele, T.; Van Camp, J. Bioaccessibility of Polyphenols from Plant-Processing Byproducts of Black Carrot (Daucus carota L.). J. Agric. Food Chem. 2016, 64, 2450–2458. [Google Scholar] [CrossRef]
- Kammerer, D.; Carle, R.; Schieber, A. Quantification of anthocyanins in black carrot extracts (Daucus carota ssp. sativus var. atrorubens Alef.) and evaluation of their color properties. Eur. Food Res. Technol. 2004, 219, 479–486. [Google Scholar] [CrossRef]
- Koley, T.K.; Singh, S.; Khemariya, P.; Sarkar, A.; Kaur, C.; Chaurasia, S.N.S.; Naik, P.S. Evaluation of bioactive properties of Indian carrot (Daucus carota L.): A chemometric approach. Food Res. Int. 2014, 60, 76–85. [Google Scholar] [CrossRef]
- Stintzing, F.C.; Stintzing, A.S.; Carle, R.; Frei, B.; Wrolstad, R.E. Color and Antioxidant Properties of Cyanidin-Based Anthocyanin Pigments. J. Agric. Food Chem. 2002, 50, 6172–6181. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Huang, Y.; Wang, F.; Song, X.; Wang, G.L.; Xiong, A.S. Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol. 2014, 14, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yildiz, M.; Willis, D.K.; Cavagnaro, P.F.; Iorizzo, M.; Abak, K.; Simon, P.W. Expression and mapping of anthocyanin biosynthesis genes in carrot. Theor. Appl. Genet. 2013, 126, 1689–1702. [Google Scholar] [CrossRef]
- Alfenito, M.R.; Souer, E.; Goodman, C.D.; Buell, R.; Mol, J.; Koes, R.; Walbot, V. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent Glutathione S-Transferases. Plant Cell 1998, 10, 1135–1149. [Google Scholar] [CrossRef] [Green Version]
- Klein, M.; Burla, B.; Martinoia, E. The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. J. FEBS Lett. 2006, 580, 1112–1122. [Google Scholar] [CrossRef] [Green Version]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef] [Green Version]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Curtin, C.; Bézier, A.; Franco, C.; Zhang, W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 2008, 59, 3621–3634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Cone, K.C.; Burr, F.A.; Burr, B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 1986, 83, 9631–9635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red coloration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [Green Version]
- Rinaldo, A.R.; Cavallini, E.; Jia, Y.; Moss, S.M.A.; McDavid, D.A.J.; Hooper, L.C.; Robinson, S.P.; Tornielli, G.B.; Zenoni, S.; Ford, C.M.; et al. A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 2015, 169, 1897–1916. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Sun, S.S.; Gugger, P.F.; Wang, Q.F.; Chen, J.M. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus (Nelumbo Adans.). PeerJ Life Environ. 2016, 4, e2369. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit coloration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.M.M.L.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Haruta, K.S.; Pitaksutheepong, C.; Abe, Y.; Kakizaki, Y.; Yamamoto, K.; Shimada, N.; Yamamura, S.; Nishihara, M. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 2008, 49, 1818–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Kimura, S.; Demura, T.; Takeda, J.; Ozeki, Y. DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol. Biol. 2005, 59, 739–752. [Google Scholar] [CrossRef]
- Wako, T.; Kimura, S.; Chikagawa, Y.; Ozeki, Y. Characterization of MYB proteins acting as transcriptional regulatory factors for carrot phenylalanine ammonia-lyase gene (DcPAL3). Plant Biotechnol. 2010, 27, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Feng, K.; Que, F.; Wang, F.; Xiong, A. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Iorizzo, M.; Cavagnaro, P.F.; Bostan, H.; Zhao, Y.; Zhang, J.; Simon, P.W. A cluster of MYB transcription factors regulates anthocyanin biosynthesis in carrot (Daucus carota L.) root and petiole. Front. Plant Sci. 2018, 9, 1927. [Google Scholar] [CrossRef] [Green Version]
- Giusti, M.; Wrolstad, R.E. Characterization and Measurement of Anthocyanins by UV-visible Spectroscopy. In Handbook of Food Analytical Chemistry; Wiley: Hoboken, NJ, USA, 2001; pp. 19–31. [Google Scholar]
- Smeriglio, A.; Denaro, M.; Barreca, D.; D’Angelo, V.; Germanò, M.P.; Trombetta, D. Polyphenolic profile and biological activities of black carrot crude extract (Daucus carota L. ssp. sativus var. atrorubens Alef.). Fitoterapia 2018, 124, 49–57. [Google Scholar] [CrossRef]
- Hirner, A.A.; Seitz, H.U. Isoforms of chalcone synthase in Daucus carota L. and their differential expression in organs from the European wild carrot and in ultraviolet-A-irradiated cell cultures. Planta 2000, 210, 993–998. [Google Scholar] [CrossRef]
- Ozeki, Y.; Davies, E. Takeda Structure and expression of chalcone synthase gene in carrot suspension cultured cells regulated by 2, 4-D. Plant Cell Physiol. 1993, 34, 1029–1037. [Google Scholar]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that color our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, S.; Akita, Y.; Ishizaka, H.; Narumi, I.; Tanaka, A. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen. J. Plant Physiol. 2012, 169, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Algarra, M.; Fernandes, A.; Mateus, N.; de Freitas, V.; Esteves da Silva, J.C.G.; Casado, J. Anthocyanin profile and antioxidant capacity of black carrots (Daucus carota L. ssp. sativus var. atrorubens Alef.) from Cuevas Bajas, Spain. Food Compos. Anal. 2014, 33, 71–76. [Google Scholar] [CrossRef]
- Gläßgen, W.E.; Rose, A.; Madlung, J.; Koch, W.; Gleitz, J.; Seitz, H.U. Regulation of enzymes involved in anthocyanin biosynthesis in carrot cell cultures in response to treatment with ultraviolet light and fungal elicitors. Planta 1998, 204, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Bannoud, F.; Ellison, S.; Paolinelli, M.; Horejsi, T.; Senalik, D.; Fanzone, M.; Iorizzo, M.; Simon, P.W.; Cavagnaro, P.F. Dissecting the genetic control of root and leaf tissue-specific anthocyanin pigmentation in carrot (Daucus carota L.). Theor. Appl. Genet. 2019, 132, 2485–2507. [Google Scholar] [CrossRef]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Ausubel, F.M.; Goodman, H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995, 8, 659–671. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, Q.; Feng, K.; Xiong, A. Changing Carrot Color: Insertions in DcMYB7 Alter the Regulation of Anthocyanin Biosynthesis and Modification. Plant Physiol. 2019, 181, 195–207. [Google Scholar] [CrossRef]
- Curaba, J.; Bostan, H.; Cavagnaro, P.F.; Senalik, D.; Mengist, M.F.; Zhao, Y.; Simon, P.W.; Iorizzo, M. Identification of an SCPL Gene Controlling Anthocyanin Acylation in Carrot (Daucus carota L.) Root. Front. Plant Sci. 2020, 10, 1770. [Google Scholar] [CrossRef]
- Chezem, W.R.; Clay, N.K. Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 2016, 131, 26–43. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Kodama, M.; Brinch-Pedersen, H.; Sharma, S.; Holme, I.B.; Joernsgaard, B.; Dzhanfezova, T.; Amby, D.B.; Vieira, F.G.; Liu, S.; Gilbert, M.T.P. Identification of transcription factor genes involved in anthocyanin biosynthesis in carrot (Daucus carota L.) using RNA-Seq. BMC Genom. 2018, 19, 811. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qi, Y.; Zhang, A.; Wu, H.; Liu, Z.; Ren, X. Molecular cloning and functional characterization of AcGST1, an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis). Plant Mol. Biol. 2019, 100, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, P.; Cheng, L. Developmental changes of carbohydrates, organic acids, amino acids, and phenolic compounds in ‘Honeycrisp’apple flesh. Food Chem. 2010, 123, 1013–1018. [Google Scholar] [CrossRef]
- Barba-Espín, G.; Glied, S.; Crocoll, C.; Dzhanfezova, T.; Joernsgaard, B.; Okkels, F.; Lütken, H.; Müller, R. Foliar-applied ethephon enhances the content of anthocyanin of black carrot roots (Daucus carota ssp. sativus var. atrorubens Alef.). BMC Plant Biol. 2017, 17, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef]





| Sample Name | CT | E | F | G | H |
|---|---|---|---|---|---|
| Raw reads | 45,469,730 | 47,158,771 | 47,299,178 | 42,815,288 | 45,142,919 |
| Clean reads | 44,276,147 | 46,187,029 | 46,303,908 | 41,744,653 | 43,245,500 |
| Clean bases | 6.64 | 6.93 | 6.945 | 6.26 | 6.49 |
| Q20 (%) | 97.595 | 97.545 | 97.575 | 97.615 | 97.7 |
| Q30 (%) | 93.625 | 93.48 | 93.56 | 93.64 | 93.675 |
| GC content | 43.9% | 44.855% | 44.86% | 44.69% | 44.715% |
| Total mapped | 28,571,418 (64.37%) |
29,128,798 (63.06%) |
28,684,092 (61.99%) |
28,468,352.5 (68.19%) |
28,480,330 (66.06%) |
| Uniquely mapped | 27,830,779.5 (62.70%) | 28,343,267 (61.36%) | 27,809,975.5 (60.09%) | 27,807,121.5 (66.61%) | 27,840,397.5 (64.58%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, G.; Clausen, S.K.; Rasmussen, S.K. Transcriptome Analysis Reveals Candidate Genes Related to Anthocyanin Biosynthesis in Different Carrot Genotypes and Tissues. Plants 2020, 9, 344. https://doi.org/10.3390/plants9030344
Meng G, Clausen SK, Rasmussen SK. Transcriptome Analysis Reveals Candidate Genes Related to Anthocyanin Biosynthesis in Different Carrot Genotypes and Tissues. Plants. 2020; 9(3):344. https://doi.org/10.3390/plants9030344
Chicago/Turabian StyleMeng, Geng, Sabine K. Clausen, and Søren K. Rasmussen. 2020. "Transcriptome Analysis Reveals Candidate Genes Related to Anthocyanin Biosynthesis in Different Carrot Genotypes and Tissues" Plants 9, no. 3: 344. https://doi.org/10.3390/plants9030344