Abstract
Background
Prostate cancer is characterized by heterogeneity in the clinical course that often does not correlate with morphologic features of the tumor. Metastasis reflects the most adverse outcome of prostate cancer, and to date there are no reliable morphologic features or serum biomarkers that can reliably predict which patients are at higher risk of developing metastatic disease. Understanding the differences in the biology of metastatic and organ confined primary tumors is essential for developing new prognostic markers and therapeutic targets.
Methods
Using Affymetrix oligonucleotide arrays, we analyzed gene expression profiles of 24 androgen-ablation resistant metastatic samples obtained from 4 patients and a previously published dataset of 64 primary prostate tumor samples. Differential gene expression was analyzed after removing potentially uninformative stromal genes, addressing the differences in cellular content between primary and metastatic tumors.
Results
The metastatic samples are highly heterogenous in expression; however, differential expression analysis shows that 415 genes are upregulated and 364 genes are downregulated at least 2 fold in every patient with metastasis. The expression profile of metastatic samples reveals changes in expression of a unique set of genes representing both the androgen ablation related pathways and other metastasis related gene networks such as cell adhesion, bone remodelling and cell cycle. The differentially expressed genes include metabolic enzymes, transcription factors such as Forkhead Box M1 (FoxM1) and cell adhesion molecules such as Osteopontin (SPP1).
Conclusion
We hypothesize that these genes have a role in the biology of metastatic disease and that they represent potential therapeutic targets for prostate cancer.
Similar content being viewed by others
Background
Prostate cancer is the most common cancer in men resulting in over 232,090 new cases and 30,350 deaths annually [1]. For prostate cancer patients, metastatic disease reflects the most adverse clinical outcome. Osseous involvement with severe bone pain and spinal cord complications occur commonly in patients with metastatic disease [2]. However there is considerable heterogeneity in outcome after primary diagnosis and currently there are no morphologic or circulating biomarkers that can accurately predict the development of metastatic disease.
Metastatic prostate cancer represents the tumor's ability to escape from the primary organ and eventually colonize a distant site. Disruption of a complex set of biological processes must occur in order for tumor cells to leave the prostate and establish themselves in a different environment. Their altered interaction with the prostate microenvironment, including the stroma and extracellular matrix, their ability to migrate into the vasculature and establish themselves in secondary organs with recruitment of vascular supply represent disruption of normal cellular processes [3]. Understanding the molecular events involved in the development of metastatic prostate cancer has the potential to identify biological determinants that can aid in prognosis and development of more effective therapies.
Using gene expression microarrays, a number of studies have characterized expression profiles of prostate cancer, normal tissue and metastatic cancers. In some cases, correlations between tumor expression signatures, clinical parameters and outcome have been identified [4–11]. Unique profiles have been reported for untreated and short-term androgen ablation treated organ-confined disease and for metastatic disease, with a subset of genes differentiating metastatic androgen ablation resistant prostate cancer (AARPC) from androgen dependent metastatic cancers [10, 12–14]. In general, metastatic prostate cancer is characterized by changes in expression of genes involved in signal transduction, cell cycle, cell adhesion, migration and mitosis. In addition to these genes, AARPCs exhibit changes in expression of the androgen receptor and enzymes involved in the sterol biosynthesis pathway [12].
Some of the genes previously reported as highly downregulated in prostate tumors may reflect the differences in cellular content of metastatic and organ-confined tissues rather than intrinsic differences in biology. In contrast with organ-confined prostate tumors which are composed of a mixture of glandular epithelial, smooth muscle and other stromal cells, metastatic tissue samples are almost exclusively epithelial, with minimal supporting stroma and absence of smooth muscle. In this study, we characterize gene expression in androgen ablation resistant metastatic tumors after removing potentially uninformative stromal genes. The deleted stromal genes consist of those reported in a recent report characterizing the gene expression patterns in the prostate stroma, tumor and normal epithelium [15]. Our results provide novel insights into the biology of metastasis.
Methods
Tumor sample procurement
All tissue samples were acquired from the Health Sciences Tissue Bank of the University of Pittsburgh Medical Center under stringent Institutional Review Board guidelines with appropriate informed consent. The 18 donor and 64 primary prostate tumor samples have been described previously [7]. Specimens were received directly from the operating room. Samples (>500 mg) were excised and snap frozen in liquid nitrogen within 30 min of excision and stored at -80°C until extraction of RNA. Metastatic tumor samples were obtained from a warm autopsy program and processed similarly to primary tumors. An H&E stained frozen section of each sample was evaluated by a pathologist, to determine epithelial and stromal content and verify the presence of tumor in the sample. Dissection of the frozen tissue block was performed with the guidance of a marked H & E slide to minimize the presence of host tissue in the metastatic samples. All samples used in the study contained >80% tumor. Metastatic tumor samples were minced and divided into two equal portions to be extracted with the sample protocol used for each set of primary tumors.
Clinical profile of cases
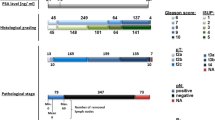
The clinical characteristics of the 64 primary tumor samples used in the Affymetrix portion of our study have been previously described [6, 7]. These cases have a mean follow-up time of 3 years. The metastatic samples consisted of 24 tissues derived from 4 patients (Table 1). All patients with metastatic disease had received androgen ablation therapy and had shown progression of disease while on androgen ablation. The clinical characteristics for the additional 10 primary prostate tumor cases used in the CodeLink study are shown in Table 1.
RNA extraction
RNA purification for the 64 primary samples has been previously described [6]. The set of metastatic samples analyzed with the Affymetrix platform was extracted with the same methodology. The set of metastatic samples and primary tumors analyzed with the CodeLink platform were extracted using the RNeasy kit (Qiagen, San Diego, CA). For the metastatic samples, one sample did not have enough for extraction with the Qiagen method, only 23 metastatic samples are included in the CodeLink assays. The concentration of each total RNA sample was measured with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies,Wilmington, DE). RNA integrity was determined by capillary electrophoresis using an Agilent 2100 Bioanalyzer (Agilent, Willmington, DE).
cRNA preparation and gene expression assays
cRNA was prepared and hybridized to Affymetrix GeneChip HGU95av2, HGU95b and HGU95c arrays (Affymetrix, Santa Clara, CA) as previously described [6]. For gene expression profiling with the CodeLink Gene Expression System (GE Healthcare, Piscataway, NJ), biotin-labeled cRNA was prepared as previously described [16]. Ten micrograms of biotin-labeled cRNA product from each sample were then fragmented with RNA fragmentation buffer at 94°C for 20 minutes. Hybridization mix was prepared according to the manufacturer's instructions and the final volume was adjusted to 260ul using nuclease-free water. The hybridization mix was heat denatured at 90°C for 5 minutes, cooled on ice and then applied to Human Uniset 20 K arrays (GE Healthcare, Piscataway, NJ). Arrays were incubated at 37°C for 18 h with shaking at 60 rpm in an Innova hybridization oven (New Brunswick, Edison, PA).
After hybridization, arrays were placed in a pre-heated (46°C) chamber filled with 0.75 × TNT (0.75 M Tris-HCL, pH 7.6, 3.75 M NaCl, Tween-20, and milli-Q water) and incubated at 46°C for 1 hour. Arrays were then stained with Streptavidin-Alexa Fluor 647 (Molecular Probes, Grand Island, NY) for 30 minutes at room temperature. Upon the completion of staining, the arrays were washed three sequential times in fresh 1 × TNT (1 M Tris-HCL, pH 7.6, 5 M NaCl, Tween-20, and milli-Q water) and then washed two final times in fresh solutions of 0.05% Tween-20 and 0.1 × SSC with gentle agitation. All arrays were dried by centrifugation at 2000 rpm for 3 minutes.
Affymetrix arrays were scanned in an Affymetrix GCS3000 Scanner (Affymetrix, Santa Clara, CA). CodeLink arrays were scanned with the GenePix 4000B scanner using GenePix Pro 4.1 software (Molecular Devices, Sunnyvale, CA).
Gene expression data analysis
The raw scanned array images from the Affymetrix GeneChip U95 arrays were processed using GCOS 1.1 software using the MAS5 algorithm (Affymetrix Corporation, Santa Clara, Ca) to generate probe cel intensity (*.cel) files. Data normalization to remove variation in overall chip intensities was performed by global scaling to a chip mean target intensity of 200 (MAS 5.0). Data for U95Av2, B and C arrays were combined for further analyses.
To identify differentially regulated genes in both datasets, these were analyzed with the Significance Analysis of Microarrays software (SAM v 1.2) [17]. Prior to analysis, genes that showed low variation across all samples were removed by using the filtering option in the Avadis 3.3 Pride Software (Strand Life Sciences, Bangalore, India) data analysis tool. To avoid false results due to difference in the tissue composition of metastatic and primary tumors, genes identified as being highly expressed in the prostatic stroma as per Stuart et al [15] were also removed. In all 1506 stromal genes and 7678 invariant genes were removed from the Affymetrix dataset. SAM generated gene lists with the lowest false discovery rates (FDR) were further analyzed for gene ontology (GO) and pathway annotations using NIH's DAVID annotation tool [18].
For CodeLink arrays, image files were analyzed with the CodeLink Expression Analysis Software version 4.1 (GE HealthCare) with use of the normalized intensity values in downstream analysis. For cross-platform comparison, Affymetrix probe sets and Codelink identifiers were mapped to Unigene ids using the DAVID annotation tool (see above). Expression data from both platforms was compared using z-transformation. Hierarchical clustering was performed using Eisen's Cluster and Treeview [19]. Data from Affymetrix experiments has been submitted to NCBI's Gene Expression Omnibus (GEO) as series GSE6919, with the following accession numbers GSE6604 (normal donor prostate), GSE6605 (metastatic prostate tumors), GSE6606 (primary prostate tumors) and GSE6608 (normal prostate tissue adjacent to tumor). Data from the CodeLink platform have been submitted to GEO with the accession number GSE6752 (primary and metastatic prostate tumors).
Quantitative real-time PCR
Differential expression of ten genes in primary and metastatic prostate cancer samples was verified with quantitative real-time PCR (QPCR) with the ABI PRISM® 7000 sequence detection system (Applied Biosystems, Foster City, CA). Three selected RNA samples from each patient were pooled together (except for patient FB666 n = 1) and therefore four RNA samples, each representing one patient, were tested. RNA samples were first heat-denatured at 70°C for 10 minutes in an Eppendorf master cycler (Eppendorf, Westbury, NY) and then chilled immediately on ice. cDNAs were reversely transcribed from one microgram of RNA using the M-MLV Reverse Transcriptase kit (Invitrogen, Carlsbad, CA), as recommended by the manufacturer. QPCR was performed based on the manufacturer's instructions with TaqMan Gene Expression Assays (Applied Biosystems) for the following genes: EGR3, SYNPO2, ANGPT2, SPP1, FOXM1, ADM, RDX, TGFBRAP1, MAK and EGR1(assay IDs: Hs00231780_m1, Hs00326493_m1, Hs01048047_mH, Hs00959010_m1, Hs00153543_m1, Hs00969450_g1, Hs00988414_g1; Hs01093285_m1; Hs01048300_m1, Hs00152928_m1). When multiple TaqMan assays for one gene were available, the assay that interrogated the sequence closest to the target sequence in the Affymetrix arrays was chosen. PCR cycles were performed according to the assay instructions in an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems). Relative quantification of the expression level of each transcript in each sample was calculated using the Delta-Delta CT method in the ABI PRISM 7000 Sequence Detection System Software (Applied Biosystems) [20]. Human reference RNA from Stratagene (Stratagene Corp., La Jolla, CA) was used as the calibrator (untreated control) and human glucuronidase beta (GUSB) gene was used as the endogenous reference gene (Forward primer: GGA ATT TTG CCG ATT TCA TGA; Reverse primer: CCG AGT GAA GAT CCC CTT TTT; Probe: 6FAM-AAC AGT CAC CGA CGA GAG TGC TGG G-TAMRA).
Results
Differential gene expression in metastatic prostate cancer and the role of stromal content in defining true downregulated genes
Differential expression analysis of the metastatic and primary tumor samples shows that a large number of the most highly downregulated genes such as TAGLN, ACTG2, TPM1, MYH111 and DES have been previously identified as expressed mostly in the prostatic stromal cells [15]. Since only the epithelial component of prostate cancer is present in metastatic tumors, this result most likely reflects the lack of stroma in metastases, and not a true down-regulation of these genes in the metastatic epithelial cells. Therefore, based on a recent report characterizing cell type specific gene expression in the prostate [15], we removed the set of genes expressed mainly by the stromal cells of the primary tumors. In all 1506 transcripts associated with a stromal signature were deleted prior to further analysis. Since the stromal genes were characterized using the U95Av2 chip and our analysis includes u95Av2, B and C chips, only stromal genes represented by probe sets on U95Av2 were removed in this modified analysis. SAM analysis shows that 1277 genes are up and 977 genes are downregulated at least 2 fold at the lowest FDR (0.01), in metastatic prostate samples (see Additional file 1). A list of the top 50 up and top 50 down regulated genes at the lowest fdr, after removing ESTs and uncharacterized clones is shown in Table 2. This list includes signal transducers, cell cycle regulators, metabolic enzymes and cell adhesion molecules. Some of the most upregulated genes in our list are EIF1AX, AR, HSPD1 and HSPCA, K-ALPHA1, MLL5, UGT2B15, and some of the most downregulated genes include WNT5B5, ANXA11, FOS and SFRP1.
Metastatic samples are heterogenous in gene expression
Using immunohistochemistry (IHC) Shah et al. have shown that metastatic samples are highly heterogenous in expression of prostate specific markers leading to the hypothesis that at the molecular level, metastatic prostate cancer may represent multiple diseases even within the same patient [21]. We examined the expression of several transcripts markers including some studied by Shah et al. and confirmed the heterogeneity of expression levels in metastatic prostate cancer tissues. Expression values in donor samples, primary and metastatic samples were compared. Prostate specific antigen (PSA/KLK3) remains high in some metastatic samples and is low or absent in others, even within the same patient (Figure 1). Interestingly, AMACR, another biomarker for prostate cancer [22] expresses a heterogenous expression pattern similar to PSA. HPN, which is overexpressed in primary cancer maintains high expression in the metastatic samples in our study. AR, while overexpressed in 23 out of the 24 metastatic samples, shows highly variable expression values in individual samples. The proto-oncogenes FOS and JUNB, which are both overexpressed in primary tumors, are consistently downregulated in all metastatic samples.
Genes regulated in all metastatic cases
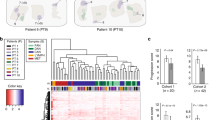
Hierarchical clustering analysis reveals that gene expression in metastatic samples is more variable between patients than between different metastatic sites from each patient (Figure 2). Although the 24 metastatic samples represent tissues from 6 metastatic sites (Table 1), no organ specific clusters were detected (Figure 2) whereas samples from the same patient tend to cluster together. Statistical comparison of organ-specific expression profiles was not attempted due to unequal distribution of samples from different metastatic sites.
Hierarchical clustering of primary and metastatic prostate cancer samples. The 24 metastatic (Mets P1, Mets P2, Mets P3 and Mets P4) and 64 primary tumor samples were clustered. The top row of color coded boxes represents metastatic or primary samples; the bottom row represents the organ from which the sample was obtained.
In order to identify probe sets that are similarly regulated in every patient, and therefore likely to represent a specific metastatic profile, the SAM differentially expressed gene list at a FDR of 2% was further filtered. For each gene on this list, a patient specific median expression value was calculated from the multiple samples from each patient. Patient P4 had only sample and this sample's signal value was considered the median value. The median values were then compared to the median value of the primary samples and those probe sets whose median value showed equal or more than a 2 fold change in every patient were considered part of the metastatic prostate cancer signature. Under this criteria 415 transcripts are upregulated and 364 are downregulated in all patients with metastasis (see Additional file 2). A truncated gene list consisting of genes regulated at least 3 fold in all patients is shown in Table 3. Upregulation of AR in all samples from metastatic cancer patients represents a known "androgen resistant" or AARPC (androgen ablation resistant prostate cancer) phenotype [12]. The transcripts identified as differentially expressed in our study exhibit similarities with a previous study of AARPC tumors [10, 12]. Cytokeratins 5 and 15 (KRT5/KRT15), markers of basal cells in prostate glands, show uniform downregulation in all metastatic tumors, confirming the absence of basal epithelial cells.
Biological annotation of differentially expressed genes in metastatic prostate cancer
The list of differentially expressed transcripts at least 2 fold in all patients was further analyzed for biological themes and gene ontology (GO) using the NIH's DAVID annotation tool. This analysis revealed that metastatic prostate cancer exhibits altered regulation of amino acid, carbohydrate and nucleotide metabolism consistent with the proliferative capacity and altered energy needs of metastatic tumors (data not shown). In the context of prostate cancer biology, genes involved in cell-adhesion, bone remodeling, cell-cycle and transcription are of particular interest (Table 4). Disruption of cell adhesion and altered interaction with the extracellular matrix is a hallmark of metastatic tumors [3]. In agreement with this, the secreted phosphoprotein and cell adhesion molecule osteopontin (SPP1) is one of the most highly upregulated transcripts in our metastatic samples. Elevated expression of SPP1 has been correlated with poor prognosis in prostate tumors and other cancers and it has often been implicated in metastasis to bone and other organs [2, 23–28]. In all 29 probe sets representing cell adhesion genes are altered in all metastatic samples. This gene list includes FN1, ITGB8, THBS2, HNT and CDH10. Genes involved in bone remodeling such as BMP4 and ANKH are also altered in expression, although none of the samples in our study are bone metastatic samples suggesting that these proteins may also be involved in cancer metastasis to other organs.
Disruption of the cell cycle is highlighted by the presence of a large number of cell-cycle related transcripts in the list of differentially expresed genes in all metastatic samples. The list contains 37 cell cycle genes, and includes SEP4, SEP7, PTN and VEGF. Similarly, a large number of transcription factors (67) including AR, SRY, FOS and EGR3, are differentially expressed. Two members of the winged-helix family of transcription factors, FoxP1 and FoxM1, show upregulation in the metastatic samples. Interestingly, FoxM1b has been shown to promote progression of prostate carcinomas in an experimental model [29].
The MAP kinase signaling pathway was also identified as being important in the metastatic process, with 26 probe sets involved in this pathway being differentially expressed in all metastatic samples. The regulated genes include DUSP1, DUSP2, DUSP8, MAP3K8, MAP4K4, FGF13, FGFR2 and FOS. Involvement of MAP kinase in androgen receptor signaling has been previously described [30].
Validation of differentially expressed transcripts with an independent set of primary tumors and different gene expression platforms confirms gene expression profiles of metastatic prostate cancer
Gene expression analysis with the CodeLink Uniset 20 K microarray was carried out for 23 of the metastatic samples and compared to an independent set of 10 primary tumors. Similar to the Affymetrix analysis (see above), hierarchical clustering of the CodeLink data set reveals heterogeneity in expression and no organ-specific clustering (data not shown). Comparison of results with the Affymetrix based dataset, based on genes with common Unigene ids on both platforms, show a similar pattern of differentially expressed genes. Of the top 1000 up and down regulated transcripts from each platform, approximately 70% share common unigene ids and of these 22% of the genes are identified as regulated by both platforms (see Additional file 3). This level of correlation is significant, given the well-documented difficulties in cross-platform comparisons of expression data [31, 32]. Examples of z-transformed expression values for selected genes in both platforms are shown in Figure 3.
Additionally, real-time quantitative PCR (QPCR) assays were performed for a selected set of genes in pooled samples for each patient with metastatic disease and 5 of the primary tumors from the CodeLink set. The transcripts for this analysis were chosen to represent diverse biological processes and were chosen from the differentially expressed genes identified as up/down-regulated in the Affymetrix/Codelink data comparison. As shown in Figure 4, qPCR assays confirmed the results from the microarray platforms. SYNPO2 and EGR3, which are downregulated and RDX and FOXM1, which are upregulated in the microarray analysis exhibit a very similar expression pattern in the qPCR analysis. Interestingly, FoxM1 is consistently upregulated in metastases, while RDX was upregulated in only two of the four patients with metastatic disease, confirming the heterogeneity of metastatic prostate cancer.
Validation of differentially expressed genes with quantitative real time PCR. QPCR was performed on RNA from samples used in the CodeLink analysis. Three selected metastatic RNA samples from each patient were pooled together (except for patient FB666 n = 1) and therefore four RNA samples, each representing one metastatic patient and 5 primary tumors were tested. The insert shows average expression values for metastatic and primary tumors.
Discussion
Despite extensive research, the molecular mechanisms of metastatic prostate cancer and androgen resistance development are still poorly understood. Our study shows that a number of biological processes including cell adhesion, cell cycle and transcription regulation are altered in metastatic disease when compared to primary tumors, and point to specific transcripts that participate in the metastatic process.
Previous investigators have reported differences in gene expression profiles of metastatic and primary prostate cancer [10, 12, 14, 21]. Our results show partial overlap with these previous characterizations of metastatic disease. Some genes that are in concordance with these studies include transcription factors such as FOXM1, and c-FOS.1. Differences in patient demographics, pathology and treatment, non-standard tissue handling, experimental and statistical methods may all contribute to differences in gene lists. Differences with other published gene lists might also reflect the fact that in our study, only samples from patients with androgen-insensitive prostate cancer were used. Additionally, in our experimental design we have incorporated features that increase the significance of our findings and increase the likelihood that the genes identified truly reflect the biology of metastatic prostate cancer. First, by subtracting transcripts previously identified as being expressed by the prostatic stroma, we have incorporated previous knowledge about the expression profiles of different components of prostate tumors in order to focus on those transcripts intrinsic to metastatic cells. This takes into account the fact that metastatic tumors do not contain all the tissue elements present in organ-confined tumors. The major benefit of this strategy is to better define the genes that are down-regulated in metastatic tumor cells. Second, by analyzing multiple tumor samples from each patient, we have addressed the fact that metastatic prostate cancer shows significant heterogeneity, even within the same patient [21]. This is corroborated by our results. and we address this issue by focusing our analysis on the transcripts that show significant differential expression in all metastatic sites within and between patients.
Multiple biological processes appear to be altered in metastatic prostate cancer. One common theme that has emerged from studies of metastatic disease is the central role of the androgen receptor in the development of androgen resistant disease. Several mechanisms including amplification of the AR gene, upregulation of mRNA expression to allow binding by low levels of androgens, mutations in the ligand binding domain (LBD) that allow the receptor to be activated by antagonists, and alteration in the normal AR signaling pathway, have been proposed to explain the ability of prostate cancer to recur in the presence of androgen ablation therapy [33, 34]. Consistent with previous observations, AR is up-regulated in all metastatic samples in our study. Similarly, gene expression changes of the MAP kinase pathway in metastasis may be related to the development of the AARPC phenotype. The gene list from our analysis shares some similarities with mouse xenograft prostate cancers models (CWR22) of androgen independence. MSMB, CCND1, EFNA3, FKBP and ADM, HGF are similarly regulated in mouse models and in our study [35–37].
Changes in the expression level of several additional transcripts may reveal clues about the mechanism of metastasis and androgen resistance. We find upregulation of the enzyme UGT2B15 in all metastatic patients. Upregulation of UGT2B15 in androgen independent prostate cancer has been reported previously [14]. This increase appears paradoxical, since UGT2B15 is involved in hormone inactivation. However, as suggested by Stanbrough et al [14], upregulation of multiple genes related with androgen metabolism might reflect that metastatic tumor cells have an increased capacity to convert weak androgens into testosterone or DHT. However, in contrast to their findings, transcripts for AKR1C3, SRD5A1, HSD3B2, AKR1C2, AKR1C1 are not consistently upregulated in the metastatic samples in our study. Interestingly, when reviewing individual values for each sample, some metastases indeed show higher levels for some of these transcripts, which might reflect the heterogeneity of metastatic prostate cancer phenotypes. Another possible explanation for this discrepancy is that the metastatic samples used by Stanbrough et al. are all from bone metastases and this type of sample is not represented in our study. Clearly, further investigation into the role of these pathway genes in the development of androgen resistance by metastatic samples is needed.
Several genes involved in cell-cell interaction and cell adhesion appear to be up-regulated in these tumors. SPP1 (osteopontin), a secreted, integrin-binding glycoprotein with adhesive properties, has been shown to be correlated with metastasis to the bone and with poor prognosis in various cancers and is highly upregulated in all the metastatic samples in our study. Elevated plasma osteopontin levels have also been correlated with lower survival and bone metastasis in hormone resistant prostate cancer [25]. Interestingly, Stanborough and collaborators also identified SPP1 as upregulated in their metastatic samples, however, their interpretation was that this increase was part of the bone response to the metastases. Our study confirms that upregulation of SPP1 is a feature intrinsic to androgen-resistant metastatic prostate cancer, independent of the site of metastasis. It has been postulated that metastasis to specific target organs may require not only expression of SPP1 but an additional set of signaling molecules that promote metastasis to the specific organ. SPP1 when expressed with IL11 has been shown to promote metastasis of breast cancer cells to the bone [38] but not to the adrenal medulla. Further detailed studies are required to address the specific role of SPP1 and other co-expressed genes in prostate cancer metastasis and whether SPP1 represents a potential therapeutic target for androgen-resistant disease. Interestingly, the gene expression profile termed as "bone module" and postulated as a hallmark of tumor metastasis to bone [39] is not dysregulated in our study, most likely reflecting the fact that we did not assay bone metastatic samples. It is also possible, that the role that SPP1 plays in metastasis to bone and/or other organs may involve distinct mechanisms [38].
Metastatic tumors have been described as undergoing an epithelial to mesenchymal transition with loss of the differentiated phenotype. Downregulation of transcription factors such as JUN has been observed in advanced stages of other cancers and its loss of activity has been postulated to be involved in this transition [40]. In our study, both FOS and JUNB, which are upregulated in primary tumors compared to normal prostate tissue are highly downregulated in the metastatic samples. FBN, also representative of the EMT transition [38] is overexpressed in our metastatic samples. Our analysis has also identified a number of additional genes, such as KLK11,STC1 and S100A8 that are uniformly regulated in all metastatic patients. The role of S100A8 in prostate cancer has been studied with evidence suggesting that it is elevated in prostate cancer and may be involved in MAP kinase and NFK-B signalling [41, 42]. STC1, involved in calcium homeostasis, has been reported to have osteoblastic and angiogenic modulator properties with altered expression in some cancers [43–45]. The serine protease KLK11 appears to be regulated in prostate cancer with negative correlation between aggressiveness and expression [46].
A recent study observed overexpression of 62 genes due to surgical manipulation related ischemia of the prostate [47]. In our study, 12 out of the 62-gene ischemia profile are downregulated in all metastatic samples. This gene list includes DUSP1, BTG2, IER2, PTGS2, NR4A1, AMD1, C20orf35, KLF4, RAB4A, KLF6, CTGF and GOLPH2. In our data set, these genes represent only 0.01% of the total number of genes differentially regulated in all metastatic samples. Since our metastatic samples all originate from autopsy studies, it is likely that they had been exposed to longer ischemia than the organ confined samples obtained from surgical specimens. Thus, if the differences we observed were related to the ischemia, we would have expected an increase in the expression of these genes, and not the observed downregulation. Therefore, it is unlikely that surgical manipulation can explain the differential gene expression between metastatic and primary tumors.
Conclusion
In summary, our results support the roles for specific cell adhesion, androgen metabolism and transcription factor genes in the development of androgen-independent metastatic prostate cancer. Furthermore, the differentially expressed transcripts in metastatic tumors that we report have been validated with two independent sets of primary tumors, two gene expression microarray platforms, and selected genes were further validated by qRT-PCR. Our results corroborate the notion that metastatic prostate cancer is quite heterogeneous within a single patient. Despite this heterogeneity our experimental design allowed us to identify common expression profiles for androgen-independent metastatic prostate cancer.
Abbreviations
- (SAM):
-
Significance Analysis of Microarray
- (FDR):
-
false discovery rate
- (AARPC):
-
androgen ablation resistant prostate cancer
References
American Cancer Society: Cancer facts and figures, 2006. 2006, Atlanta , American Cancer Society
Logothetis CJ, Lin SH: Osteoblasts in prostate cancer metastasis to bone. Nat Rev Cancer. 2005, 5 (1): 21-28. 10.1038/nrc1528.
Stewart DA, Cooper CR, Sikes RA: Changes in extracellular matrix (ECM) and ECM-associated proteins in the metastatic progression of prostate cancer. Reprod Biol Endocrinol. 2004, 2: 2-10.1186/1477-7827-2-2.
Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM: Delineation of prognostic biomarkers in prostate cancer. Nature. 2001, 412 (6849): 822-826. 10.1038/35090585.
Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M: Gene expression analysis of prostate cancers. Mol Carcinog. 2002, 33 (1): 25-35. 10.1002/mc.10018.
Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH: Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004, 22 (14): 2790-2799. 10.1200/JCO.2004.05.158.
Chandran UR, Dhir R, Ma C, Michalopoulos G, Becich M, Gilbertson J: Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005, 5 (1): 45-10.1186/1471-2407-5-45.
Chetcuti A, Margan S, Mann S, Russell P, Handelsman D, Rogers J, Dong Q: Identification of differentially expressed genes in organ-confined prostate cancer by gene expression array. Prostate. 2001, 47 (2): 132-140. 10.1002/pros.1056.
Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR: Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002, 1 (2): 203-209. 10.1016/S1535-6108(02)00030-2.
LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL: Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002, 62 (15): 4499-4506.
True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, Beer TM, Gelmann E, Datta M, Mostaghel E, Knudsen B, Lange P, Vessella R, Lin D, Hood L, Nelson PS: A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci U S A. 2006, 103 (29): 10991-10996. 10.1073/pnas.0603678103.
Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, Ryan C, Smith S, Scher H, Scardino P, Reuter V, Gerald WL: Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004, 164 (1): 217-227.
Bismar TA, Demichelis F, Riva A, Kim R, Varambally S, He L, Kutok J, Aster JC, Tang J, Kuefer R, Hofer MD, Febbo PG, Chinnaiyan AM, Rubin MA: Defining aggressive prostate cancer using a 12-gene model. Neoplasia. 2006, 8 (1): 59-68. 10.1593/neo.05664.
Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, Febbo PG, Balk SP: Increased Expression of Genes Converting Adrenal Androgens to Testosterone in Androgen-Independent Prostate Cancer. Cancer Res. 2006, 66 (5): 2815-2825. 10.1158/0008-5472.CAN-05-4000.
Stuart RO, Wachsman W, Berry CC, Wang-Rodriguez J, Wasserman L, Klacansky I, Masys D, Arden K, Goodison S, McClelland M, Wang Y, Sawyers A, Kalcheva I, Tarin D, Mercola D: In silico dissection of cell-type-associated patterns of gene expression in prostate cancer. Proc Natl Acad Sci U S A. 2004, 101 (2): 615-620. 10.1073/pnas.2536479100.
Ma C, Lyons-Weiler M, Liang W, LaFramboise W, Gilbertson JR, Becich MJ, Monzon FA: In Vitro Transcription Amplification and Labeling Methods Contribute to the Variability of Gene Expression Profiling with DNA Microarrays. J Mol Diagn. 2006, 8 (2): 183-192. 10.2353/jmoldx.2006.050077.
Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001, 98 (9): 5116-5121. 10.1073/pnas.091062498.
Eisen MB, Spellman PT, Brown PO, Botstein D: Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998, 95 (25): 14863-14868. 10.1073/pnas.95.25.14863.
Livak KJ, Schmittgen TD: Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-[Delta][Delta]CT Method. Methods. 2001, 25 (4): 402-10.1006/meth.2001.1262.
Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, Kim R, Rubin MA, Pienta KJ: Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004, 64 (24): 9209-9216. 10.1158/0008-5472.CAN-04-2442.
Zhou M, Chinnaiyan AM, Kleer CG, Lucas PC, Rubin MA: Alpha-Methylacyl-CoA racemase: a novel tumor marker over-expressed in several human cancers and their precursor lesions. Am J Surg Pathol. 2002, 26 (7): 926-931. 10.1097/00000478-200207000-00012.
Weber GF: The metastasis gene osteopontin: a candidate target for cancer therapy. Biochim Biophys Acta. 2001, 1552 (2): 61-85.
Reinholz MM, Iturria SJ, Ingle JN, Roche PC: Differential gene expression of TGF-beta family members and osteopontin in breast tumor tissue: analysis by real-time quantitative PCR. Breast Cancer Res Treat. 2002, 74 (3): 255-269. 10.1023/A:1016339120506.
Hotte SJ, Winquist EW, Stitt L, Wilson SM, Chambers AF: Plasma osteopontin: associations with survival and metastasis to bone in men with hormone-refractory prostate carcinoma. Cancer. 2002, 95 (3): 506-512. 10.1002/cncr.10709.
Wai PY, Kuo PC: The role of Osteopontin in tumor metastasis. J Surg Res. 2004, 121 (2): 228-241. 10.1016/j.jss.2004.03.028.
Furger KA, Menon RK, Tuck AB, Bramwell VH, Chambers AF: The functional and clinical roles of osteopontin in cancer and metastasis. Curr Mol Med. 2001, 1 (5): 621-632. 10.2174/1566524013363339.
Forootan SS, Foster CS, Aachi VR, Adamson J, Smith PH, Lin K, Ke Y: Prognostic significance of osteopontin expression in human prostate cancer. Int J Cancer. 2006, 118 (9): 2255-2261. 10.1002/ijc.21619.
Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, Lyubimov A, Costa RH: Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006, 66 (3): 1712-1720. 10.1158/0008-5472.CAN-05-3138.
Shimada K, Nakamura M, Ishida E, Konishi N: Molecular roles of MAP kinases and FADD phosphorylation in prostate cancer. Histol Histopathol. 2006, 21 (4): 415-422.
Hollingshead D, Lewis DA, Mirnics K: Platform influence on DNA microarray data in postmortem brain research. Neurobiol Dis. 2005, 18 (3): 649-655. 10.1016/j.nbd.2004.10.020.
Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P: Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005, 33 (18): 5914-5923. 10.1093/nar/gki890.
Setlur SR, Rubin MA: Current thoughts on the role of the androgen receptor and prostate cancer progression. Adv Anat Pathol. 2005, 12 (5): 265-270. 10.1097/01.pap.0000184179.50672.f7.
Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL: Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004, 10 (1): 33-39. 10.1038/nm972.
Sirotnak FM, She Y, Khokhar NZ, Hayes P, Gerald W, Scher HI: Microarray analysis of prostate cancer progression to reduced androgen dependence: studies in unique models contrasts early and late molecular events. Mol Carcinog. 2004, 41 (3): 150-163. 10.1002/mc.20051.
Amler LC, Agus DB, LeDuc C, Sapinoso ML, Fox WD, Kern S, Lee D, Wang V, Leysens M, Higgins B, Martin J, Gerald W, Dracopoli N, Cordon-Cardo C, Scher HI, Hampton GM: Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen-independent prostate cancer xenograft model CWR22-R1. Cancer Res. 2000, 60 (21): 6134-6141.
Mousses S, Bubendorf L, Wagner U, Hostetter G, Kononen J, Cornelison R, Goldberger N, Elkahloun AG, Willi N, Koivisto P, Ferhle W, Raffeld M, Sauter G, Kallioniemi OP: Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res. 2002, 62 (5): 1256-1260.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J: A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003, 3 (6): 537-549. 10.1016/S1535-6108(03)00132-6.
Segal E, Friedman N, Koller D, Regev A: A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004, 36 (10): 1090-1098.
Hulboy DL, Matrisian LM, Crawford HC: Loss of JunB activity enhances stromelysin 1 expression in a model of the epithelial-to-mesenchymal transition of mouse skin tumors. Mol Cell Biol. 2001, 21 (16): 5478-5487. 10.1128/MCB.21.16.5478-5487.2001.
Hermani A, De Servi B, Medunjanin S, Tessier PA, Mayer D: S100A8 and S100A9 activate MAP kinase and NF-kappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp Cell Res. 2006, 312 (2): 184-197. 10.1016/j.yexcr.2005.10.013.
Hermani A, Hess J, De Servi B, Medunjanin S, Grobholz R, Trojan L, Angel P, Mayer D: Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005, 11 (14): 5146-5152. 10.1158/1078-0432.CCR-05-0352.
Zlot C, Ingle G, Hongo J, Yang S, Sheng Z, Schwall R, Paoni N, Wang F, Peale FV, Gerritsen ME: Stanniocalcin 1 is an autocrine modulator of endothelial angiogenic responses to hepatocyte growth factor. J Biol Chem. 2003, 278 (48): 47654-47659. 10.1074/jbc.M301353200.
McCudden CR, Majewski A, Chakrabarti S, Wagner GF: Co-localization of stanniocalcin-1 ligand and receptor in human breast carcinomas. Mol Cell Endocrinol. 2004, 213 (2): 167-172. 10.1016/j.mce.2003.10.042.
Chirgwin JM, Mohammad KS, Guise TA: Tumor-bone cellular interactions in skeletal metastases. J Musculoskelet Neuronal Interact. 2004, 4 (3): 308-318.
Stavropoulou P, Gregorakis AK, Plebani M, Scorilas A: Expression analysis and prognostic significance of human kallikrein 11 in prostate cancer. Clin Chim Acta. 2005, 357 (2): 190-195. 10.1016/j.cccn.2005.03.026.
Lin DW, Coleman IM, Hawley S, Dumpit R, Gifford D, Kezele P, Hung H, Knudsen BS, Kristal AR, Nelson PS: Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. J Clin Oncol. 2006, 24 (23): 3763-3770. 10.1200/JCO.2005.05.1458.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/7/64/prepub
Acknowledgements
This work was funded by NCI/NIH grant 5 U01 CA88110-02 (GM) "Molecular Reclassification of Prostate Cancer" Director's Challenge for the Molecular Classification of Cancer Consortium; by the Pennsylvania Cancer Alliance Bioinformatics Consortium (PCABC) Grant from the PA Department of Health ME-01740 (MJB) and a College of American Pathologists Foundation Scholar's Award to FAM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
URC was involved in data analysis, results interpretation and manuscript preparation. RD was involved in the histologic evaluation of samples. CM performed the QPCR experiments and was involved with data analysis and results interpretation. MLW and WJL performed the Affymetrix and CodeLink experiments. GM and MB participated in conceptualization and study design and manuscript review. FM was responsible for general oversight of the study, providing technical direction, guidance for the analysis team, and participated in manuscript preparation. All authors have read and approved the final manuscript.
Electronic supplementary material
12885_2006_714_MOESM1_ESM.xls
Additional file 1: Differentially expressed genes between metastatic and primary prostate tumors. Results of SAM analysis of the 24 metastatic and 64 primary tumors. The Affymetrix probe set id, gene names and assignment of biological process for each gene is shown. (XLS 286 KB)
12885_2006_714_MOESM2_ESM.xls
Additional file 2: Comparison of median expression values of all samples from each metastatic patient with primary tumor expression values. After SAM analysis of the 25 metastatic and 64 primary tumors, genes whose median expression values differ at two in each metastatic patient compared to the median value of primary tumor samples were selected. (XLS 153 KB)
12885_2006_714_MOESM3_ESM.xls
Additional file 3: Genes identified as regulated in metastatic prostate cancer from both Codelink and Affymetrix platforms (XLS 50 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chandran, U.R., Ma, C., Dhir, R. et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 7, 64 (2007). https://doi.org/10.1186/1471-2407-7-64
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-7-64