OrthoFinder Tutorials Phylogenetic orthology inference for comparative genomics
OrthoFinder
OrthoFinder is a software program for phylogenetic orthology inference.
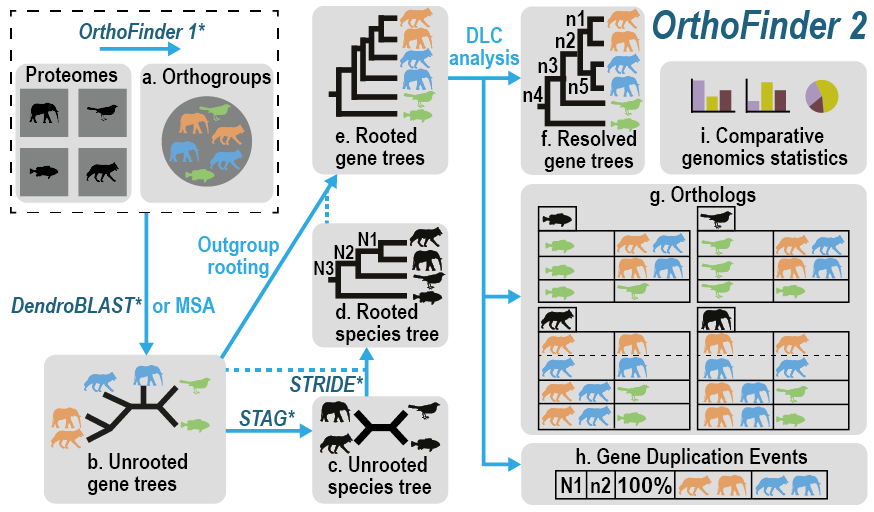
It takes as input the proteomes for the species you want to analyse and from these it automatically:
- infers the orthogroups for your species
- infers a complete set of rooted gene trees
- infers a rooted species tree
- infers all orthology relationships between the genes using the gene trees
- infers gene duplication events and cross references them to the corresponding nodes on the gene and species trees
- provides comparative genomics statistics for your species
As well as large scale analyses, it can be used to carefully check individual ortholog relationships prior to experimental studies.
OrthoFinder vs OrthoMCL and other orthology inference software
Unlike other orthology inference software OrthoFinder uses gene trees. This means you can check every ortholog relationship in the gene tree it came from. The use of gene trees gives very high ortholog inference accuracy:
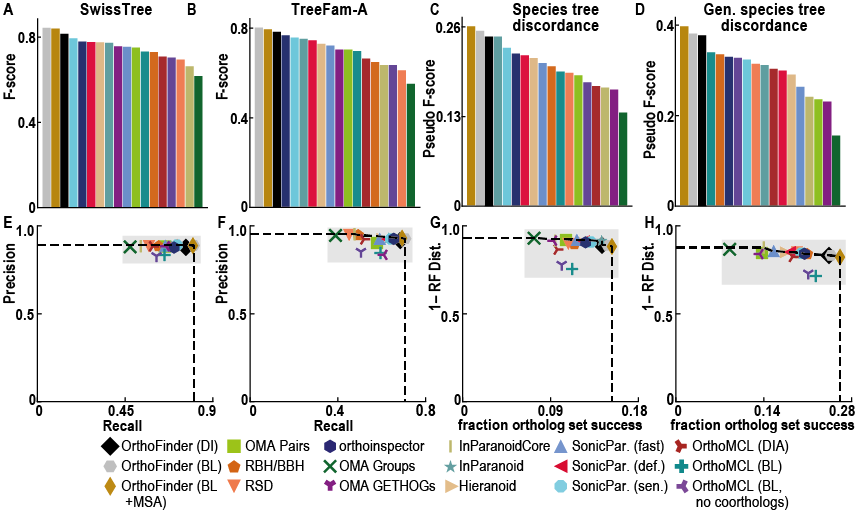
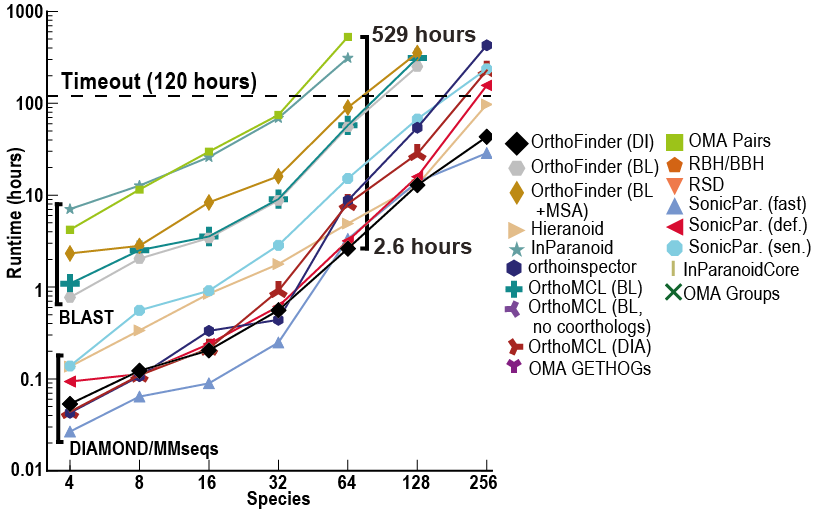
Despite OrthoFinder using a more rigorous, gene-tree based approach to ortholog inference it is incredibly fast! In addition to this, it also returns far more comparative genomics data than other methods (orthogroups, orthologs, rooted genes trees, gene duplication events etc.)