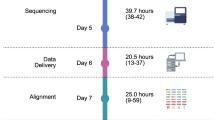
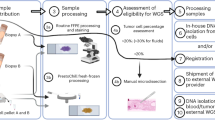
Abstract
Pancreatic cancer is a highly lethal malignancy that presents multiple technical challenges for genomic studies. Next-generation sequencing and its applications have proven successful in the study of other tumour types, unravelling the interplay between DNA and RNA changes that are unique to the tumour. This Review outlines the genomic studies performed to date that have explored the somatic alterations of pancreatic cancer genomes, setting the stage for the introduction of our current technological capabilities. In spite of several challenging aspects posed by pancreatic tumours in particular and clinical sequencing-based diagnostics in general, next-generation sequencing and analysis can now be used in experiments relating to the treatment of patients with this disease. As a means to improve patient outcomes, the application of comprehensive next-generation sequencing and analysis to the genomes of patients with pancreatic cancer to identify therapeutic options is proposed.
Key Points
Pancreatic cancer presents multiple challenges to cancer genomic studies, although early studies have defined the predominant mutations in the disease
New approaches, based on next-generation sequencing and analysis, might overcome some of these challenges and provide new information about tumour heterogeneity and the full mutational spectrum of each patient's disease
Medical interpretation of sequencing data from DNA isolated from pancreatic tumours and matched normal tissue, and RNA isolated from pancreatic tumours, by integrated analysis might identify therapeutic options among small-molecule or antibody-based inhibitors
Diagnostic trials that combine next-generation sequencing and medical interpretation of data with the use of targeted therapies should be designed for testing in patients with pancreatic cancer
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
National Cancer Institute. Surveillance Epidemiology and End Results [online] (2012).
Koorstra, J. B., Hustinx, S. R., Offerhaus, G. J. & Maitra, A. Pancreatic carcinogenesis. Pancreatology 8, 110–125 (2008).
Ghaneh, P., Costello, E. & Neoptolemos, J. P. Biology and management of pancreatic cancer. Gut 56, 1134–1152 (2007).
Jemal, A. et al. Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 (2009).
Fisher, W. E. The promise of a personalized genomic approach to pancreatic cancer and why targeted therapies have missed the mark. World J. Surg. 35, 1766–1769 (2011).
Rowley, J. D., Golomb, H. M. & Dougherty, C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukaemia. Lancet 1, 549–550 (1977).
Rowley, J. D. et al. Further evidence for a non-random chromosomal abnormality in acute promyelocytic leukemia. Int. J. Cancer 20, 869–872 (1977).
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Mardis, E. R. A decade's perspective on DNA sequencing technology. Nature 470, 198–203 (2011).
Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008).
Welch, J. S. et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA 305, 1577–1584 (2011).
Roychowdhury, S. et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci. Transl. Med. 3, 111ra121 (2011).
The Genome Reference Consortium [online], (2012).
1000 Genomes. A deep catalog of human genetic variation [online] (2012).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Mardis, E. R. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9, 387–402 (2008).
Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012).
Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6, 677–681 (2009).
Wang, J. et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods 8, 652–654 (2011).
Koboldt, D. C. et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Albert, T. J. et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods 4, 903–905 (2007).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 39, 1522–1527 (2007).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Myers, R. M. et al. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9, e1001046 (2011).
Iorio, M. V. & Croce, C. M. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4, 143–159 (2012).
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009).
International Cancer Genome Consortium. ICGC cancer genome projects [online] (2012).
National Cancer Institute. The cancer genome atlas [online] (2012).
International Cancer Genome Consortium. Pancreatic cancer—ductal adenocarcinoma [online] (2011).
International Cancer Genome Consortium. Rare pancreatic tumors—enteropancreatic endocrine tumors and rare pancreatic exocrine tumors [online] (2010).
Jones, S. J. et al. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 11, R82 (2010).
Forbes, S. A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 (2011).
Wellcome Trust Sanger Institute. Catalogue of somatic mutations in cancer [online] (2012).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Gonzalez-Perez, A. & Lopez-Bigas, N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 88, 440–449 (2011).
Dummer, R. & Flaherty, K. T. Resistance patterns with tyrosine kinase inhibitors in melanoma: new insights. Curr. Opin. Oncol. 24, 150–154 (2012).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Quach, N., Goodman, M. F. & Shibata, D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin. Pathol. 4, 1 (2004).
Wilson, R. K. et al. Mutational profiling in the human genome. Cold Spring Harb. Symp. Quant. Biol. 68, 23–29 (2003).
Pao, W. et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Campbell, P. J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010).
Haeno, H. et al. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell 148, 362–375 (2012).
Hsu, C. C. et al. Adjuvant chemoradiation for pancreatic adenocarcinoma: the Johns Hopkins Hospital-Mayo Clinic collaborative study. Ann. Surg. Oncol. 17, 981–990 (2010).
Artinyan, A., Anaya, D. A., McKenzie, S., Ellenhorn, J. D. & Kim, J. Neoadjuvant therapy is associated with improved survival in resectable pancreatic adenocarcinoma. Cancer 117, 2044–2049 (2011).
Mann, K. M. et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl Acad. Sci. USA 109, 5934–5941 (2012).
Olive, K. P. et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 324, 1457–1461 (2009).
Kraman, M. et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 330, 827–830 (2010).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Mardis, E. Applying next-generation sequencing to pancreatic cancer treatment. Nat Rev Gastroenterol Hepatol 9, 477–486 (2012). https://doi.org/10.1038/nrgastro.2012.126
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2012.126
This article is cited by
Induction of cancer neoantigens facilitates development of clinically relevant models for the study of pancreatic cancer immunobiology
Cancer Immunology, Immunotherapy (2023)
Ein Fall von Pankreaskarzinom
Forum (2018)
Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance
Journal of Experimental & Clinical Cancer Research (2013)
Personal Genomic Measurements: The Opportunity for Information Integration
Clinical Pharmacology & Therapeutics (2013)
A new method for the determination of critical polyethylene glycol concentration for selective precipitation of DNA fragments
Applied Microbiology and Biotechnology (2013)