-
PDF
- Split View
-
Views
-
Cite
Cite
Astrid Vabret, Julia Dina, Stéphanie Gouarin, Joëlle Petitjean, Sandrine Corbet, François Freymuth, Detection of the New Human Coronavirus HKU1: A Report of 6 Cases, Clinical Infectious Diseases, Volume 42, Issue 5, 1 March 2006, Pages 634–639, https://doi.org/10.1086/500136
Close - Share Icon Share
Abstract
Background. Human coronavirus HKU1 (HCoV-HKU1), a new group 2 coronavirus, was first characterized in 2005 from 2 adults with pneumonia in Hong Kong, China. To the best of our knowledge, there is no other report to date about the detection of this new virus. We report a molecular method allowing for the detection of HCoV-HKU1 and also report the clinical presentation of 6 infected patients.
Methods. We screened 141 specimens (135 nasal samples and 6 stool samples) received in February and March 2005 in our laboratory and obtained from 135 hospitalized patients (61.5% of whom were <5 years old and 34.1% of whom were >20 years old) for HCoV-HKU1.
Results. HCoV-HKU1 was detected in 6 (4.4%) of the 135 nasal specimens and in 2 (33.3%) of the 6 stool samples; the positive samples were obtained from 6 patients (5 children and 1 adult). The clinical presentation of these 6 patients was as follows: 3 were admitted to the hospital for acute enteric disease resulting in severe dehydration associated with upper respiratory symptoms; 1 had fever, otitis, and febrile seizure; 1 had a sample obtained to investigate failure to thrive; and 1 had a sample obtained for exploration of X-linked agammaglobulinemia and hyperleucocytosis.
Conclusion. HCoV-HKU1 can be detected in respiratory and stool samples from children and adults in a part of the world other than Hong Kong. Our results suggest that HCoV-HKU1 could be associated with respiratory and enteric diseases, and its detection can be related to a persistent asymptomatic infection in patients with poor underlying conditions.
Coronaviruses (family Coronaviridae, order Nidovirales) are enveloped viruses with a linear, nonsegmented, positive-sense, single-stranded RNA genome and are divided into 3 distinct groups (group 1, group 2, and group 3). Five types of human coronaviruses (HCoV) have been described: HCoV-OC43 and HCoV-229E have been recognized since the mid-1960s and belong to group 1 and group 2. In 2003, the severe acute respiratory syndrome (SARS)—associated coronavirus (SARS-CoV) was identified, but it has not yet been assigned to one of the groups [1]. In 2004, another group 1 coronavirus, HCoV-NL63, was reported in The Netherlands [2, 3]. In January 2005, a new group 2 coronavirus, HCoV-HKU1, was found in 2 patients with pneumonia in Hong Kong. This new coronavirus was detected by RT-PCR of the pol gene of coronaviruses with use of conserved primers in the nasopharyngeal aspirates of patients. The complete genome was sequenced, and its general organization concurs with those of other coronaviruses. It contains the HE gene, which characterizes the group 2 coronaviruses, and has a very intriguing feature consisting of a variable number of tandem copies of a 30-bp repeat region that codes for a highly acidic domain (ATR) in ORF1a (nsp1 region). Despite the high number of cell cultures inoculated with respiratory samples, HCoV-HKU1 could not be recovered from cell culture [4]. No cytopathic effect was observed, and RT-PCR performed on the culture supernatant and cell lysates showed a lack of viral replication. Isolating HCoV on cell culture is very difficult. HCoV prototype strains 229E, OC43, and NL63 can replicate in MRC5 cells, HRT18 cells, and LLC-MK2 cells, respectively, but there are still few primary isolates. Woo et al. [4] reported that SARS-CoV can be recovered only from <20% of patients infected with SARS-CoV. There is no other report to date concerning the detection of HCoV-HKU1. In our laboratory, we conducted a preliminary study of 135 respiratory samples to know if this new coronavirus can be detected in a part of the world other than Hong Kong. The purpose of this article is to present the method used for the detection of HCoV-HKU1 and report the clinical presentation associated with this infection.
Materials And Methods
We screened arbitrarily for HCoV-HKU1 135 HUH7 cell culture showing an extensive lysis at day 4 after onset of infection. HUH7 cells were inoculated with respiratory samples collected from 135 patients hospitalized in February and March 2005 at the University Hospital of Caen (Caen, France). The age distribution of the tested patients was as follows: 83 patients (61.5%) were <5 years of age, 6 patients (4.4%) were 6–20 years of age, and 46 patients (34.1%) were >21 years of age. The tested respiratory specimens were obtained from patients with respiratory symptoms and then were sent to the laboratory for viral diagnosis. The patients or, in the case of minors, the parents or legal guardians of the patients consented to having their samples tested for respiratory viruses, including coronaviruses. The samples were processed routinely for immunofluorescence direct antigen assay using monoclonal antibodies against influenza A and B viruses; respiratory syncytial virus; parainfluenzavirus types 1, 2, and 3; and adenovirus (Imagen). All samples had negative results. It is part of our strategy to inoculate samples with negative results onto a cell culture system (HUH7 cell line) [5]. In this protocol, HuH7 cells were grown in RPMI-1640 medium with glutamine with 5% fetal calf serum (Gibco; Invitrogen), penicillin (100 U/mL), and gentamycin (50 µg/mL), and were distributed in 48-well tissue culture microplates. When HUH7 cells were 80% confluent, the growth medium was discarded and each well was inoculated with 100 µL of the clinical samples. Microplates were centrifugated at 1500 rpm for 30 min at 30°C. Samples were then removed, and each well was filled with 1 mL of the same culture medium supplemented with 2% fetal calf serum (Gibco; Invitrogen). By day 4 after onset of infection, HuH7 cells were examined for cytopathic effect and the supernatant were used for amplification techniques. Cytopathic effect characteristics are as follows: small cells, no refringent, cells scattered on the culture, and a progressive lysis of the cell sheet. The RNA was extracted using the High pure RNA isolation kit (Roche) and tested with RT-PCR techniques previously described and used for the detection of rhinovirus, enterovirus, influenza virus C, HCoV-229E, HCoV-OC43, and HCoV-NL63 [6, 7]. A 1-step HCoV-HKU1 RT-PCR amplification of a 443-bp fragment was performed using the OneStep RT-PCR kit (Qiagen) and the following primers set defined in N gene from the GenBank sequence database (accession number AY597011): HKU1 sens (5′-ACCAATCTGAGCGAAATTACCAAAC-3′) and HKU1 antisense (5′-CGGAAACCTAGTAGGGATAGCTT-3′). The reaction was undertaken using OneStep RT-PCR kit (Qiagen, Courtaboeuf) according to the manufacturer's protocol. Each RT-PCR test was performed using the usual precautions to avoid contamination. No cross-reaction of this RT-PCR assay was observed with the other classical HCoV. The assay was performed without a positive control. RT-PCR products were processed on an agarose gel stained with ethidium bromide and visualized under UV light. The positive results were confirmed by sequencing from the amplified product and by RT-PCR performed directly with use of the respiratory specimens. For 2 patients with results positive for detection of HCoV-HKU1 in respiratory specimens, stool samples obtained at the same time were available and have been tested. Two other RT-PCR assays were performed using the specimen that was positive for HCoV-HKU1 detection, one of which involved amplification of a 713-bp fragment with S1 gene—specific primers HKU1-S1 sens (5′-ACCACAGTTCCTCGCATAAGT-3′) and HKU1-S1 antisense (5′-AGATATTGGCGTTTAGAC-3′), the other amplifying a variable size fragment with primers defining on both sides of the highly acidic domain in ORF1a (ATR-HKU1 sens: 5′-AATGGCCTCTCGTATGTAT-3′; ATR-HKU1 antisense: 5′-TTACAAGTAACACAGAACGCA-3′). The RT-PCR products were sequenced, and the obtained nucleotide sequences of the partial S1 gene were compared with the prototype strain sequences available in GenBank. The alignments were prepared using Clustal X, version 1.83. A phylogenetic tree was constructed by the neighbor-joining method, and bootstrap values were determined by 1000 replicates. We used HCoV-OC43 as an outgroup. The medical records were examined retrospectively for the 6 patients with test results positive for HCoV-HKU1.
Results
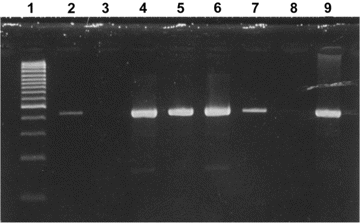
Among the 135 cell cultures tested with RT-PCR, the positive samples consisted of 10 coronaviruses (6 positive for HCoV-HKU1, 2 positive for HCoV-NL63, and 2 positive for HCoV-OC43), 23 rhinoviruses, 1 enterovirus, and 1 influenza virus C. There were 2 codetections, one of which consisted of HCoV-HKU1 and influenza virus C, and the other of which consisted of rhinovirus and HCoV-OC43. To eliminate cell culture contamination, the N gene HCoV-HKU1 RT-PCR was performed directly on the respiratory specimens obtained from the 6 patients. All of these specimens, as well as the stool samples available for 2 of these patients, were found to have positive results. The 443-bp amplicon resulting from the amplification of the partial N gene was sequenced for 6 of the 8 specimens with positive results (GenBank accession numbers DQ131635-DQ131641) (figure 1). Our isolates shared 98%–100% nucleotide identity with the same region of the HCoV-HKU1 reference strain. We then sequenced the 713-bp amplicon corresponding to partial spike gene of 4 positive specimens. Genetic sequences from other positive specimens could not be obtained, because the primers used for the amplification of the partial spike gene did not amplify a PCR product of sufficient quantity to allow for reliable genetic sequencing analysis. The phylogenetic analysis of 603-bp of these sequences (GenBank accession numbers DQ131642-DQ131645) shows that our isolates form a group containing sequences with different markers and that there is no cross-contamination between the different isolates. Three of these isolates clustered with the prototype strain described in Hong Kong. One isolate (DQ131645) had characteristics of an outlier (figure 2). To determine if our isolates contained a variable number of tandem copies of 30-bp repeat region—the significant feature of HCoV-HKU1—we sequenced amplicon resulting from a RT-PCR of the region of ORF1a. To confirm the number of ATRs, this test was conducted several times for 2 isolates. The results show that there were 7 and 8 ATRs in the 2 isolates tested, compared with 11 and 14 ATRs recorded in the 2 Hong Kong isolates [4].
Ethidium bromide stain of 2% agarose gel showing RT-PCR products (443 bp) of partial coronavirus N gene with human coronavirus HKU1—specific primers. Lane 1, size marker (100 bp); lanes 2, 4–7, and 9, positive respiratory samples; lanes 3 and 8, negative control RT-PCR mix.
Phylogenetic analysis of a 603-bp region of partial spike gene of 4 human coronavirus (HCoV)-HKU1 isolates from France. The phylogenetic tree was constructed by the neighbor-joining method, and bootstrap values were determined by 1000 replicates in Clustal X, version 1.83. To construct the tree, the reference strain HCoV-HKU1 (NC006577) was included. HCoV-OC43 is used as the outgroup.
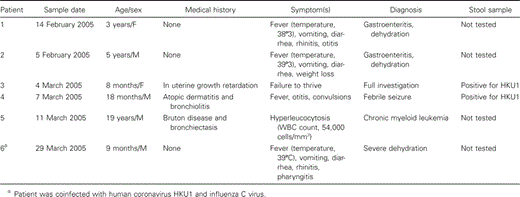
HCoV-HKU1 was detected in nasal and stool samples obtained from 6 patients, 4 of whom were male, and 2 of whom were female (table 1). The age of the infected patients ranged from 8 months to 5 years, with the exception of one 19-year-old man (patient 5). The age distribution of the patients infected with HCoV-HKU1 and the age distribution of tested patients were closed. The medical records of all 6 patients were examined. Three of them (patients 1, 2, and 6) were admitted to the hospital for severe dehydration due to fever, vomiting, and diarrhea. Upper respiratory symptoms (rhinitis, pharyngitis, or otitis) were also reported. Unfortunately, no stool samples for these patients were available in the hospital. Patient 4 had a febrile seizure and otitis. Both respiratory and stool samples tested positive for HCoV-HKU1. Patients 3 and 5 were admitted to the hospital for a complete investigation and did not suffer from acute symptoms; patient 3 was an 8-month-old girl with failure to thrive, and patient 5 was a 19-year-old man with Bruton agammaglobulinemia and hyperleucocytosis with a final diagnosis of chronic myeloid leukaemia. A stool sample obtained from patient 3 was available and tested positive for HCoV-HKU1.
Clinical presentation of patients infected with human coronavirus HKU1.
Discussion
HCoV-HKU1 was first characterized in Hong Kong in January 2005 in the respiratory specimens of 2 adults with pneumonia. HCoV-HKU1 is a previously unrecognized group 2 coronavirus. The clinical significance of this viral infection was made evident in the index patient by the combined evolution of clinical symptoms, viral loads in the respiratory samples, and development of a specific antibody response. This index patient was a 71-year-old Chinese man with poor underlying conditions (pulmonary tuberculosis complicated by bronchiectasis, a history of being a chronic smoker, and obstructive airway disease). He was admitted to the hospital for pneumonia and was discharged after 5 days of hospitalization. However, respiratory, stool, and urine samples were collected from him weekly for a 5-week period. The viral load in respiratory specimens decreased during the second week of the illness and was undetectable in the third week. The decrease in viral load was accompanied by recovery from the illness and development of a specific antibody response. HCoV-HKU1 was detected only in respiratory specimens, not in stool and urine samples [4]. There is no other report to date about the geographic and temporal distribution and the clinical symptoms associated with this virus.
In our study, the arbitrary decision was made to screen 135 HUH7 cell cultures showing a cytopathic effect at day 4 after onset of infection and inoculated by nasal specimens collected during 2 winter months, February and March 2005. The choice of HUH7 cells was made because some studies showed that these cells appear to have a broad spectrum, allowing for the detection of a wide range of viruses, including coronaviruses 229E, OC43, and MHV [8–10]. Unfortunately, we failed to amplify HCoV-HKU1 on HUH7 cells by serial passage. At first passage, we could see a cytopathic effect on day 4. However, because of the lack of specific antibodies against HCoV-HKU1, we could not test HUH7 cells to know whether there were infected cells showing viral replication. So, the HCoV-HKU1 RNA detected by RT-PCR may be a residual virus from a clinical sample or a virus amplified in cell culture in the first passage on HUH7 cells. The duration of incubation is limited to 4 days, which may be insufficient for the growth of coronaviruses. More experiments are being conducted to adapt HKU1 strains to cell line. Six patients were infected with HCoV-HKU1, including 5 children and 1 adult. Because samples were obtained mainly from pediatric departments, 61.5% of the tested patients were children. So, the high frequency with which HCoV-HKU1 was detected in children was biased because of the recruitment of the samples. There is no evidence that children are more commonly infected by HCoV-HKU1 than other individuals. Positive specimens were collected throughout the study period, from 14 February to 24 March 2005. This distribution did not indicate an outbreak. HCoV strains OC43, 229E, and NL63 are reported to have a winter seasonality in temperate regions [2, 8, 9, 10]. Our study only suggests that HCoV-HKU1 could also have a winter seasonality in these regions. A better understanding of the epidemiology of different HCoV in different parts of the world could contribute to better management of the molecular and serological diagnosis of future emerging or reemerging viruses, such as SARS-CoV. Because of the lack of a systematized selection of the tested samples, the patients with positive results are presented as case reports.
It should be noted that one-half of the patients were admitted to the hospital not for respiratory illnesses, but for gastroenteritis resulting in severe dehydration associated with upper respiratory symptoms. In effect, previous studies have suggested that coronaviruses other than SARS-CoV may be involved in enteric diseases, despite the fact that there is no clear evidence that they cause enteric illness [9, 11–13]. This would not be surprising in view of the clear involvement of SARS-CoV and some animal coronavirus strains in severe diarrheal diseases. We have shown that HCoV-HKU1 can be detected in stool samples. Nevertheless, further studies must be conducted to confirm the potential dual tropism (respiratory and enteric) of this group 2 coronavirus.
HCoV-HKU1 was also detected in 2 patients without acute symptoms and with poor underlying conditions (patients 3 and 5). Although HCoV (except SARS-CoV) were generally associated with acute and self-limiting respiratory infections, this detection of viral RNA can be related to a persistent or asymptomatic chronic infection. Chiu et al. [14] reported a repeated detection of HCoV-229E in an immunocompromised child >3 months old. The genetic analysis of a 305-bp region of the spike protein region of 229E revealed that these 3 specimens had identical genetic sequences, providing evidence of virus persistence rather than reinfection. Patient 6, who tested positive for HCoV-HKU1, had an influenza C virus detected in the same clinical specimen. It is not possible to determine whether this is true coinfection or whether 1 of the viruses represents continued shedding from a previous infection. It is interesting to note that the genome of group 2 coronaviruses and influenza C virus both contain the HE gene. Phylogenetic analysis suggested that the HE genes of coronaviruses and influenza C have a common ancestral origin. Because HCoV and influenza C virus infect similar tissues, the significant sequence homology between the HE genes of the 2 viruses suggests that coinfection followed by recombination could have occurred in the past [4, 15]. The expression of the HE gene has been shown to be heterogeneous in different species of group 2 coronaviruses. The biological significance of HE protein is not well understood. Further experiments have to be performed to determine the essentiality and function of HE in HCoV-HKU1, especially in cell tropism.
In conclusion, we report a molecular method allowing for the detection of the new coronavirus HKU1. We are able to detect HKU1 in respiratory samples and stool specimens, indicating that this virus can be excreted in this way. Our results indicate that HCoV-HKU1 is present in hospitalized patients with mild respiratory and enteric tract illness. Determining whether or not HCoV-HKU1 can cause enteric diseases will require further investigations. Phylogenetic analysis of our isolates suggest that both variants of HCoV-HKU1 may cocirculate. Ours is the first observation and report of HCoV-HKU1 infection in France. Future studies will yield more-accurate data about the circulation of this virus.
Acknowledgments
Potential conflicts of interest. All authors: no conflicts.
Financial support. The European Commission EPISARS contract (SP22-CT-2004-511063) and the Programme de Recherche en Réseaux Franco-Chinois (épidémie du SRAS: de l'émergence au contrôle).






Comments