-
PDF
- Split View
-
Views
-
Cite
Cite
Erick Antezana, Mikel Egaña, Bernard De Baets, Martin Kuiper, Vladimir Mironov, ONTO-PERL: An API for supporting the development and analysis of bio-ontologies, Bioinformatics, Volume 24, Issue 6, March 2008, Pages 885–887, https://doi.org/10.1093/bioinformatics/btn042
Close - Share Icon Share
Abstract
Motivation: Many biomedical ontologies use OBO or OWL as knowledge representation language. The rapid increase of such ontologies calls for adequate tools to facilitate their use. In particular, there is a pressing need to programmatically deal with such ontologies in many applications, including data integration, text mining, as well as semantic applications supporting translational research.
Results: We present an Application Programming Interface (API) called ONTO-PERL. This API significantly extends the repertoire of available tools supporting the development and analysis of bio-ontologies.
Availability: The source code code as well as sample usage scripts can be found at: http://search.cpan.org/dist/ONTO-PERL/
Contact: erick.antezana@psb.ugent.be
1 INTRODUCTION
Ontologies support consistent and unambiguous knowledge sharing and provide a framework for knowledge integration. More specifically, ontologies represent the agreed knowledge about a domain of discourse. The knowledge is represented by creating a single model with the terms of the domain as well as the relationships between those terms (Stevens et al., 2007). The relationships between terms effectively define what properties a given term must have. Entities are also linked to human readable information like labels. Thus, an ontology links term labels to their interpretations, i.e. specifications of their meanings, defined as a set of properties. As such, ontologies can be used to support automatic semantic interpretation of textual information, thereby providing a basis for advanced text mining (Doms et al., 2005; Müller et al., 2004). Moreover, structured and integrated knowledge provides a basis for advanced reasoning to validate hypotheses and generate new knowledge (Blake et al., 2006; Myhre et al., 2006). Reasoning services can be used to re-engineer the design of parts of the whole ontology (such as classification) or to design entirely new extensions that comply with the current knowledge (Wolstencroft et al., 2007). All these scenarios and applications need foundational tools to deal with ontologies.
OBO1 and OWL2 are becoming the de facto knowledge representation languages in the biomedical domain. OBO is human readable and it has gained wide acceptance. Many ontologies, such as GO (The Gene Ontology Consortium, 2000), are expressed in OBO. However, OBO does not have an explicit and well-defined semantics. In contrast, OWL is computer readable since it does have such a semantics, and, hence, automated reasoning can be performed on OWL ontologies.
Several tools are currently available to manage and develop OBO and OWL ontologies, either in the form of ontology editors or APIs. Within the bio-ontology community, OBO-Edit Day-Richter07 (OBO-centered) and Protégé3 (OWL-centered) are the most frequently used ontology-building environments. Protégé also has a plug-in for loading OBO ontologies (Moreira et al., 2007). Both ontology editors offer open java APIs that can be used to build applications and explore bio-ontologies. There also exist some independent APIs (or API-like tools) in java and perl. In java, OWL or OBO ontologies can be loaded and managed with the OWL API.4 In PERL, go-perl,5 GO::Term::Finder (Boyle et al., 2004) and Bio::Ontology6 are available. go-perl and GO::Term::Finder are GO-specific, and therefore many bio-ontologies, such as those under the OBO foundry,7 cannot be handled easily without tweaking the code. Bio::Ontology is not GO-specific but it lacks important functionalities, for instance, to intersect two ontologies, unify ontologies, export to different formats (OWL, XML, DOT, etc). Moreover, it lacks modularity in annotations (such as def, synonym and dbxref). Therefore, we present ONTO-PERL, an OBO-centered PERL API that provides a turnkey service to help bio-ontologists handle ontologies, do data exploration and perform mining.
2 IMPLEMENTATION
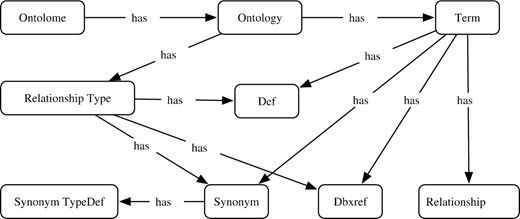
ONTO-PERL comprises an extensible set of object-oriented PERL modules that can be used for programmatically working with ontologies. ONTO-PERL can be installed as any typical CPAN module.8 A set of comprehensive test files is included in the distribution. The object model is strongly influenced by the OBO language specification [versions 1.0 and 1.2 (refer to footnote no. 1)]. Therefore, there is basically one PERL module per atomic OBO entity: Term, Relationship, Def, Synonym, Dbxref, IDspace, Ontology and SynonymTypeDef. Figure 1 depicts a simplified object architecture.
ONTO-PERL provides a set of features right out of the box. First, it has an organized set of subroutines and structures for dealing with ontologies. Second, ONTO-PERL is not tied to any operating system. Third, the model behind the ontology structure is fully compatible with the current OBO specification (v1.2) so that any ontology in OBO format can be parsed and then easily manipulated in an object-oriented manner.
Some modules included in the standard PERL distribution are required to enable some of the functionalities available in ONTO-PERL. For example, XML::Simple needs to be installed to convert OBO files into OWL files (and vice versa), according to the oboInOwl mapping.9
ONTO-PERL is the subject of intensive ongoing development. It already supports a rich set of features for ontology building. It can be integrated easily into any PERL application or any other supporting PERL modules. It offers many interfaces for dealing with ontologies in general, e.g. two or more ontologies can be merged (given an identical idspace), sub-ontologies can be retrieved as well as children terms of a given term. Table 1 shows some types of operations that can be executed with ONTO-PERL. Finally, conversion utilities are also available for having ontologies in OBO, DOT,10 XML GML,11 RDF,12 or OWL format for diverse applications (e.g. querying, visual exploration, reasoning).
Sample operations that can be executed with ONTO-PERL
| No. . | Operation . |
|---|---|
| 1 | Find all the terms and/or relationships in a given ontology o |
| 2 | Retrieve all the descendants of a given term T |
| 3 | Retrieve all the ancestors of a given term T |
| 4 | Find the intersection of two given ontologies o1 and o2 |
| 5 | Find the terms by synonym or alternate label |
| 6 | List the terms that are obsolete |
| 7 | List all the terms with a given database reference |
| 8 | Find out the total number of terms and relationships |
| 9 | Merge two given ontologies o1 and o2 |
| 10 | Get a sub-ontology from a given ontology o |
| 11 | Find the path(s) between term T1 and term T2 |
| No. . | Operation . |
|---|---|
| 1 | Find all the terms and/or relationships in a given ontology o |
| 2 | Retrieve all the descendants of a given term T |
| 3 | Retrieve all the ancestors of a given term T |
| 4 | Find the intersection of two given ontologies o1 and o2 |
| 5 | Find the terms by synonym or alternate label |
| 6 | List the terms that are obsolete |
| 7 | List all the terms with a given database reference |
| 8 | Find out the total number of terms and relationships |
| 9 | Merge two given ontologies o1 and o2 |
| 10 | Get a sub-ontology from a given ontology o |
| 11 | Find the path(s) between term T1 and term T2 |
Sample operations that can be executed with ONTO-PERL
| No. . | Operation . |
|---|---|
| 1 | Find all the terms and/or relationships in a given ontology o |
| 2 | Retrieve all the descendants of a given term T |
| 3 | Retrieve all the ancestors of a given term T |
| 4 | Find the intersection of two given ontologies o1 and o2 |
| 5 | Find the terms by synonym or alternate label |
| 6 | List the terms that are obsolete |
| 7 | List all the terms with a given database reference |
| 8 | Find out the total number of terms and relationships |
| 9 | Merge two given ontologies o1 and o2 |
| 10 | Get a sub-ontology from a given ontology o |
| 11 | Find the path(s) between term T1 and term T2 |
| No. . | Operation . |
|---|---|
| 1 | Find all the terms and/or relationships in a given ontology o |
| 2 | Retrieve all the descendants of a given term T |
| 3 | Retrieve all the ancestors of a given term T |
| 4 | Find the intersection of two given ontologies o1 and o2 |
| 5 | Find the terms by synonym or alternate label |
| 6 | List the terms that are obsolete |
| 7 | List all the terms with a given database reference |
| 8 | Find out the total number of terms and relationships |
| 9 | Merge two given ontologies o1 and o2 |
| 10 | Get a sub-ontology from a given ontology o |
| 11 | Find the path(s) between term T1 and term T2 |
3 RESULTS AND DISCUSSION
Systems biology projects increasingly require the integration of a range of ontology-driven integrated solutions including genomic data, proteomic data and modeling facilities that enable hypothesis generation. These so-called mashup systems usually need a sound building environment. ONTO-PERL addresses and eases the ontology-related aspects. ONTO-PERL has been successfully used to build an automatic data integration pipeline for the Cell-Cycle Ontology (CCO) (Antezana et al., 2006). Many sample applications are included in the ONTO-PERL distribution. The most interesting ones include parsers for specific data, such as NCBI taxonomy,13 UniProt,14 and IntAct.15 Although these applications are CCO specific, they can be adapted very easily to any ontology.
Some ontology providers offer their ontologies per se without appropriate tools for enabling, for instance, exploratory data analysis. Bio-ontologists therefore experience a growing need for tools (such as APIs) that support analysis or ontology engineering. The design aspects of ONTO-PERL have been carefully revised several times to optimize ease of use, features, documentation and so on. Moreover, ONTO-PERL ensures a stable behavior so that it could be part of critical tasks or be included in big software architectures that might be time-consuming to adapt. Finally, the design also considered issues to allow the API to evolve easily over time.
ACKNOWLEDGEMENTS
Research was funded by EU FP6 (LSHG-CT-2004-512143). M.E. was funded by EU FP6 Marie Curie EST (MEST-CT-2004-414632). We also thank the users community for providing valuable feedback.
Conflict of Interest: none declared.
http://search.cpan.org/dist/ONTO-PERL/ (PERL license)
REFERENCES
Author notes
Associate Editor: Alex Bateman