-
PDF
- Split View
-
Views
-
Cite
Cite
Jaime E. Blair, S. Blair Hedges, Molecular Phylogeny and Divergence Times of Deuterostome Animals, Molecular Biology and Evolution, Volume 22, Issue 11, November 2005, Pages 2275–2284, https://doi.org/10.1093/molbev/msi225
Close - Share Icon Share
Abstract
The phylogenetic relationships among deuterostome animals have been debated for many years, and a diversity of hypotheses have been proposed based on both morphological and molecular data. Here we have assembled sequences of 217 nuclear-encoded proteins to address specific questions concerning their relationships and times of origin. We recovered significant support for urochordates as the closest relative of vertebrates with an analysis of 59 proteins (17,400 amino acids) and suggest that the basal position of urochordates found in previous molecular studies may have been the result of long-branch attraction biases. Our results also support Ambulacraria, the pairing of hemichordates with echinoderms (nine proteins; 2,382 amino acids), and Cyclostomata, the pairing of lampreys with hagfish (25 proteins; 6,895 amino acids). In addition, 325 shared proteins (102,110 amino acids) were obtained from the complete genomes of six vertebrates and a urochordate for phylogenetic analysis and divergence time estimation. An evolutionary timescale was estimated using a local (Bayesian) molecular clock method. We found that most major lineages of deuterostomes arose prior to the Cambrian Explosion of fossils (∼520 MYA) and that several lineages had originated before periods of global glaciation in the Precambrian.
Introduction
Deuterostomes are traditionally distinguished from other bilaterian animals (i.e., protostomes) by a number of developmental features, including initial cleavage planes, the fate of the blastopore, and the origin of the mesoderm during embryogenesis. Exceptions to these defining characteristics have been pointed out and classic affinities questioned (e.g., Lovtrup 1975; Field et al. 1988). Five major groups are usually recognized within Deuterostomia at the level of phylum: hemichordates, echinoderms, cephalochordates, urochordates, and vertebrates. Other groups such as pogonophorans and chaetognaths, once considered deuterostomes (Barnes 1968), have been allied with protostomes based on molecular data (Telford and Holland 1993; Wada and Satoh 1994; Winnepenninckx, Backeljau, and De Wachter 1995). Despite intensive study, some major aspects of the deuterostome phylogeny remain unresolved, including the relationships among chordates and between tetrapods and the lobe-finned fish (Sarcopterygii). In addition, the quality of the fossil record of deuterostomes varies among phyla. Calcified skeletons in vertebrates and echinoderms may have increased the fossilization potential of these two groups compared to hemichordates or cephalochordates. The chitinous tunic of some urochordates has also left evidence in the fossil record for the antiquity of this lineage (e.g., Chen et al. 2003).
Within the deuterostomes, vertebrates, cephalochordates, and urochordates are usually allied as chordates (Chordata) (R. C. Brusca and G. J. Brusca 1990; Nielsen 1995). A close relationship between vertebrates and cephalochordates, to the exclusion of urochordates, has traditionally been supported by morphology (e.g., Maisey 1986). Molecular data have also recovered a vertebrate-cephalochordate group (Turbeville, Schulz, and Raff 1994; Wada and Satoh 1994; Cameron, Garey, and Swalla 2000; Winchell et al. 2002), but more recently, some studies have suggested a closer relationship between vertebrates and urochordates based on “total-evidence” trees combining morphological and molecular data (Zrzavy et al. 1998; Giribet et al. 2000), gene structure (Oda et al. 2002), and multiple protein alignments (Philippe, Lartillot, and Brinkmann 2005). Interpretations of enigmatic fossils, the cornutes and mitrates, as stem- and crown-group chordates also led Jefferies to suggest a closer relationship between vertebrates and urochordates (Jefferies 1986). Urochordates tend to have long branches in molecular phylogenies, raising the possibility of long-branch attraction as a biasing factor on their phylogenetic position.
The remaining deuterostome phyla, hemichordates and echinoderms, are considered to be closely related, and this pairing has been termed Ambulacraria (Metschnikoff 1869). Previously, hemichordates were thought to be paraphyletic, with enteropneusts (acorn worms) more closely related to chordates and pterobranchs closer to echinoderms (Barnes 1968; Nielsen 1995). Differences in larval and adult morphologies may have contributed to the confusion as tornaria larval characteristics have grouped hemichordates and echinoderms (e.g., Hyman 1959), while adult pharyngeal clefts and dorsal nerve cords have allied hemichordates with chordates (e.g., Barnes 1968). More recently, molecular data have shown that hemichordates are monophyletic and that pterobranchs may, in fact, be derived from within enteropneusts (Halanych 1995; Cameron, Garey, and Swalla 2000; Winchell et al. 2002). A close relationship between hemichordates and echinoderms has been supported with total-evidence trees (Turbeville, Schulz, and Raff 1994; Zrzavy et al. 1998; Giribet et al. 2000), molecular sequence data (Wada and Satoh 1994; Halanych 1995; Castresana et al. 1998; Bromham and Degnan 1999; Cameron, Garey, and Swalla 2000; Furlong and Holland 2002; Winchell et al. 2002), and similarities in Hox gene complements (Peterson 2004). In addition, a simple marine worm, Xenoturbella bocki, has been allied with hemichordates and echinoderms using molecular data (Bourlat et al. 2003). Within echinoderms, the relationships between the five classes (starfish, sea urchins, brittle stars, sea cucumbers, and sea lilies) has not been resolved (Littlewood et al. 1997; Janies 2001), but the consensus view places sea lilies (crinoids) as the most basal group.
While the monophyly of vertebrates has not been questioned, relationships within the phylum have received much attention. Especially controversial has been determining the closest relative of tetrapods, which has important implications for theories on the conquest of land. Fossil evidence suggests that an extinct group of lobe-finned fish, the elpistostegalids, is the closest relative of tetrapods (Ahlberg and Milner 1994; Long and Gordon 2004). Most molecular phylogenies have agreed in implicating a lobe-finned fish as the closest relative, but there has been no consensus as to the correct relationship among tetrapods and the two extant lineages (lungfish and the coelacanth) based on either morphological (e.g., Rosen et al. 1981; Forey 1986; Northcutt 1986; Schultze 1986) or molecular evidence (Meyer and Wilson 1990; Stock et al. 1991; Meyer and Dolven 1992; Hedges, Hass, and Maxson 1993; Yokobori et al. 1994; Zardoya and Meyer 1996, 1997; Cao et al. 1998; Zardoya et al. 1998; Tohyama et al. 2000; Venkatesh, Erdmann, and Brenner 2001; Brinkmann et al. 2004a, 2004b; Takezaki et al. 2004).
Among the jawed fish (Gnathostomata), cartilaginous fish (Chondrichthyes) are assumed to be a monophyletic group closely related to bony fish (Osteichthyes) (but see Martin 2001). An unorthodox scenario placing cartilaginous fish within bony fish (“Pisces”) has been suggested (Rasmussen and Arnason 1999a, 1999b; Arnason, Gullberg, and Janke 2001; Arnason et al. 2004) but has received no additional support. Jawless fish (Agnatha), including lampreys and hagfish, are widely recognized as the closest living relatives of gnathostomes (Romer 1966). Morphological data have previously suggested that lampreys are more closely related to gnathostomes (Lovtrup 1977; Hardisty 1979; Maisey 1986), a claim supported by some molecular data (Suzuki et al. 1995; Rasmussen, Janke, and Arnason 1998). Most molecular studies, however, have supported a closer relationship between lampreys and hagfish (Stock and Whitt 1992; Mallatt and Sullivan 1998; Kuraku et al. 1999; Suga et al. 1999; Hedges 2001; Mallatt, Sullivan, and Winchell 2001; Delarbre et al. 2002; Takezaki et al. 2003), which together form the Cyclostomata.
In addition to these phylogenetic uncertainties, a central question in deuterostome evolution concerns the times of origin of the major lineages and their relationship to the Cambrian Explosion of fossils. Previous molecular clock studies have suggested Precambrian origins for deuterostomes and other metazoans (Runnegar 1982; Wray, Levinton, and Shapiro 1996; Feng, Cho, and Doolittle 1997; Gu 1998; Wang, Kumar, and Hedges 1999; Nei, Xu, and Glazko 2001; Hedges et al. 2004), although little evidence exists in the fossil record prior to the Cambrian Explosion (∼520 MYA). Recent discoveries of Precambrian fossil embryos (Chen et al. 2000) and small bilaterians (Chen et al. 2004) have been dated to <580 MYA (Condon et al. 2005). Here, we have assembled available nuclear-encoded protein sequence data along with complete genome sequences to address phylogenetic relationships among deuterostomes. Molecular divergence times were also estimated using a Bayesian local clock method, which allows rates to vary among lineages, and robust fossil information to define minimum constraints on each node.
Methods
Protein sequence data were obtained from the public database (National Center for Biotechnology Information Entrez) for deuterostome and out-group taxa; a protostome (typically Drosophila) was used as a known out-group to deuterostomes. In the rare case when a protostome sequence was unavailable (<2% of proteins), a more distant out-group, either a cnidarian or a fungus, was used. Mitochondrial-encoded and short (<100 amino acids) proteins were excluded. Orthology was assessed for each protein using reciprocal Blast best hits (Altschul et al. 1997) and by visually inspecting preliminary trees for monophyly of known groups (e.g., mammals, vertebrates, echinoderms). A total of 217 proteins that individually address some aspect of deuterostome phylogeny were obtained and analyzed during this stage. Individual protein data sets were aligned (Thompson et al. 1997), and the shape parameter of the gamma distribution was estimated to model rate variation (Yang 1997). Subsets of the total 217 proteins were then concatenated to address specific relationships and were analyzed using neighbor joining (NJ) (Kumar, Tamura, and Nei 2004) and maximum likelihood (ML) (Strimmer and von Haeseler 1996) under a Jones Taylor-Thornton (JTT) + gamma model, and maximum parsimony (MP) branch and bound tree searches (Kumar, Tamura, and Nei 2004). Bootstrapping was used to determine confidence values for each node (2,000 replicates for NJ and MP and 1,000 puzzling steps for ML). Concatenations did not include any missing data (i.e., all taxa were present for all proteins). Alternative topologies were tested using an ML test (Yang 1997; Shimodaira and Hasegawa 1999) and P values (P(SH)) were recorded. A list of individual proteins used in each concatenation is included in the Supplementary Data (Supplementary Table 1, Supplementary Material online), and all protein alignments are available from the author (J.E.B.).
Bayesian Divergence Time Estimates (MYA) for Deuterostome Taxa
|
Nodea . |
Number of Proteins . |
Fossil Minimum Timeb . |
Divergence Timec . |
|---|---|---|---|
| a | 325 | 310 (370) | 326 (311, 354) |
| b | 191 | 340 (370) | 370d |
| c | 48 | 390 | 430 (421, 438) |
| d | 325 | 425 (495) | 476 (442, 494) |
| e | 65 | 457 | 525 (494, 580) |
| f | 25 | 325 | 520 (461, 596) |
| g | 51 | 520 | 652 (605, 742) |
| h | 325 | 520 | 794 (685, 918) |
| i | 75 | 520 | 891 (810, 1,067) |
| j | 19 | 485 | 581 (500, 669) |
| k | 3 | 520 | 730e |
| l | 9 | 520 | 876 (725, 1,074) |
| m
|
71
|
520
|
896 (832, 1,022)
|
|
Nodea . |
Number of Proteins . |
Fossil Minimum Timeb . |
Divergence Timec . |
|---|---|---|---|
| a | 325 | 310 (370) | 326 (311, 354) |
| b | 191 | 340 (370) | 370d |
| c | 48 | 390 | 430 (421, 438) |
| d | 325 | 425 (495) | 476 (442, 494) |
| e | 65 | 457 | 525 (494, 580) |
| f | 25 | 325 | 520 (461, 596) |
| g | 51 | 520 | 652 (605, 742) |
| h | 325 | 520 | 794 (685, 918) |
| i | 75 | 520 | 891 (810, 1,067) |
| j | 19 | 485 | 581 (500, 669) |
| k | 3 | 520 | 730e |
| l | 9 | 520 | 876 (725, 1,074) |
| m
|
71
|
520
|
896 (832, 1,022)
|
Nodes as presented in figure 4.
Fossil times used as minimum (maximum in parentheses) constraints are described in Methods.
Mean of the posterior distribution and the 95% credibility interval are shown for each divergence.
The divergence time between amniotes and amphibians approached the maximum constraint of 370 MYA (divergence time without constraint, 392 [386–398] MYA).
The divergence between sea lilies and eleutherozoans (starfish + sea urchins here) is presented as the midpoint between nodes j and l because of the low number of proteins available for time estimation.
Bayesian Divergence Time Estimates (MYA) for Deuterostome Taxa
|
Nodea . |
Number of Proteins . |
Fossil Minimum Timeb . |
Divergence Timec . |
|---|---|---|---|
| a | 325 | 310 (370) | 326 (311, 354) |
| b | 191 | 340 (370) | 370d |
| c | 48 | 390 | 430 (421, 438) |
| d | 325 | 425 (495) | 476 (442, 494) |
| e | 65 | 457 | 525 (494, 580) |
| f | 25 | 325 | 520 (461, 596) |
| g | 51 | 520 | 652 (605, 742) |
| h | 325 | 520 | 794 (685, 918) |
| i | 75 | 520 | 891 (810, 1,067) |
| j | 19 | 485 | 581 (500, 669) |
| k | 3 | 520 | 730e |
| l | 9 | 520 | 876 (725, 1,074) |
| m
|
71
|
520
|
896 (832, 1,022)
|
|
Nodea . |
Number of Proteins . |
Fossil Minimum Timeb . |
Divergence Timec . |
|---|---|---|---|
| a | 325 | 310 (370) | 326 (311, 354) |
| b | 191 | 340 (370) | 370d |
| c | 48 | 390 | 430 (421, 438) |
| d | 325 | 425 (495) | 476 (442, 494) |
| e | 65 | 457 | 525 (494, 580) |
| f | 25 | 325 | 520 (461, 596) |
| g | 51 | 520 | 652 (605, 742) |
| h | 325 | 520 | 794 (685, 918) |
| i | 75 | 520 | 891 (810, 1,067) |
| j | 19 | 485 | 581 (500, 669) |
| k | 3 | 520 | 730e |
| l | 9 | 520 | 876 (725, 1,074) |
| m
|
71
|
520
|
896 (832, 1,022)
|
Nodes as presented in figure 4.
Fossil times used as minimum (maximum in parentheses) constraints are described in Methods.
Mean of the posterior distribution and the 95% credibility interval are shown for each divergence.
The divergence time between amniotes and amphibians approached the maximum constraint of 370 MYA (divergence time without constraint, 392 [386–398] MYA).
The divergence between sea lilies and eleutherozoans (starfish + sea urchins here) is presented as the midpoint between nodes j and l because of the low number of proteins available for time estimation.
Complete genome sequences were obtained for human (version 35.1) (Lander et al. 2001; Venter et al. 2001), Mus musculus (version 33b.1) (Waterston et al. 2002), Rattus norvegicus (version 3d.1) (Gibbs et al. 2004), the domestic fowl Gallus gallus (version 1.1a) (Hillier et al. 2004), the pufferfish Takifugu rubripes (version 2c.1) (Aparicio et al. 2002) and Tetraodon nigroviridis (version 1a.1) (Jaillon et al. 2004), and the tunicate Ciona intestinalis (version 1.0) (Dehal et al. 2002). Complete genome transcripts were obtained from the Ensembl Genome Browser (http://www.ensembl.org/index.html) for all taxa except Ciona, which was obtained from the Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). Those proteins shared among taxa and present in a single copy (panorthologs) within each genome were determined (Blair, Shah, and Hedges 2005). Pairwise orthologs were first identified between genomes and then assembled into three-member clusters; those clusters containing a single sequence per taxon were retained and further intersected with other three-member clusters until all genomes were compared. Sequences from the Drosophila melanogaster genome (version 3b.1) (Adams et al. 2000) were added to each cluster to serve as the out-group. A total of 325 shared proteins were assembled from the intersection of seven deuterostome genomes; individual data sets were aligned and concatenated as described above.
Divergence times were estimated for concatenated data sets using a Bayesian local clock analysis (Kishino, Thorne, and Bruno 2001) with the following fossil times as minimum constraints: mouse-rat 12 MYA (Jacobs and Downs 1994), primate-rodent 65 MYA (Benton 2000), mammal-bird 310 MYA (maximum constraint 370 MYA) (Benton 2000), lamprey-hagfish 325 MYA (Donoghue, Smith, and Sansom 2004), amniote-amphibian 340 MYA (maximum constraint 370 MYA) (Paton, Smithson, and Clack 1999; Ruta and Coates 2004), tetrapod-lobe-finned fish 390 MYA (Benton 1990), tetrapod-ray-finned fish (Actinoptergyii) 425 MYA (maximum constraint 495 MYA) (Donoghue, Smith, and Sansom 2004), bony fish-cartilaginous fish 457 MYA (Donoghue, Smith, and Sansom 2004), starfish-sea urchins 485 MYA (Smith 1988), eleutheroids (starfish + sea urchins)-sea lilies 520 MYA (Smith 1988), agnathans-gnathostomes 520 MYA (Shu et al. 1999), vertebrates-cephalochordates 520 MYA (Shu, Conway Morris, and Zhang 1996), and vertebrates-urochordates 520 MYA (Chen et al. 2003). As noted, maximum constraints were imposed on three divergences within vertebrates. While the use of maximum constraints may cause molecular divergence times to be underestimated (Hedges et al. 1996; Hedges and Kumar 2004), these constraints are considered to be conservative based on paleontological arguments. The presence of Late Devonian transitional fossils supports the use of 370 MYA as a maximum constraint for the amniote-amphibian divergence (Benton 2000; Clack 2002). Also, the presence of stem-group gnathostomes supports a maximum age of 495 MYA for the origin of ray-finned fish (Donoghue, Forey, and Aldridge 2000; Donoghue, Smith, and Sansom 2004). At least one maximum constraint is usually necessary in a Bayesian time estimation analysis; removing all maximum constraints caused divergence times to increase by over 50%, a result clearly inconsistent with other aspects of the vertebrate fossil record.
Mean values for the prior distribution of the rate parameter (RtRate in Supplementary Table 1, Supplementary Material online) were estimated for each data set by calculating the average in-group branch length (JTT model) and dividing by the depth of the in-group node; out-groups were excluded during timing analyses, and therefore, the use of protostomes as a known out-group to deuterostomes did not affect divergence time estimates. Divergence times were also estimated under a multigene global clock (Hedges and Kumar 2003), using a Poisson + gamma model and the mammal-bird split (310 MYA) for rate calibration.
Results and Discussion
Phylogenetic Analyses
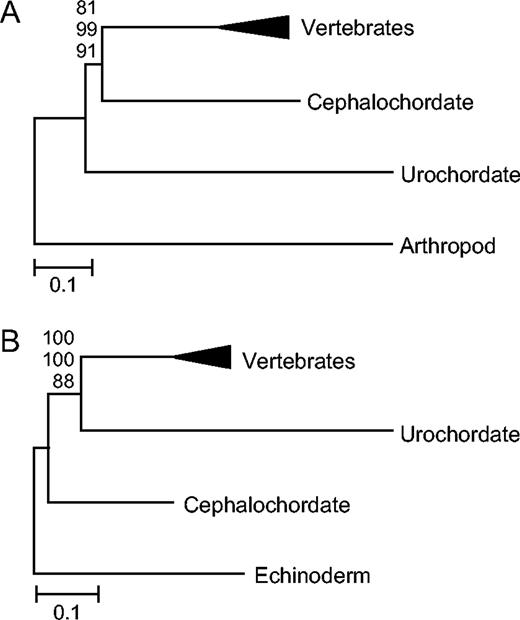
Subsets of the 217 proteins obtained from the public database were concatenated to address specific phylogenetic questions within deuterostomes. Special attention was given to the relationships among chordates. Previous molecular studies have supported a close relationship between vertebrates and cephalochordates (Turbeville, Schulz, and Raff 1994; Wada and Satoh 1994; Cameron, Garey, and Swalla 2000; Winchell et al. 2002), a group generally suggested by morphological data (Maisey 1986). Conversely, some recent studies have proposed that urochordates should be considered the closest relative of vertebrates (Zrzavy et al. 1998; Giribet et al. 2000; Oda et al. 2002; Philippe, Lartillot, and Brinkmann 2005). With a concatenation of 59 proteins (17,400 amino acids), cephalochordates were significantly placed as the closest relative of vertebrates (fig. 1A). However, the urochordate branch was approximately 50% longer than the vertebrate and cephalochordate branches, making its basal position suspect. The log likelihoods of the two possibilities (vertebrates + urochordates and vertebrates + cephalochordates) also were not significantly different (−178,100.7 vs. −178,121.1 ± 26, P(SH) = 0.23).
Phylogenetic relationships among chordate phyla. (A) The traditional relationship among chordates showing the long branch of urochordates (59 proteins, α = 0.58). (B) The influence of using echinoderms as the out-group (19 proteins, α = 0.56). NJ topologies using a JTT + gamma model are shown. Bootstrap values for NJ (top), ML (middle), and MP (bottom) are shown above each node.
When an echinoderm was used to root the chordate phylogeny (19 proteins; 4,877 amino acids), the position of urochordates changed to become the closest relative of vertebrates with strong bootstrap support (fig. 1B), although the urochordate branch was approximately twice as long as the vertebrate and cephalochordate branches. An ML test of this data set significantly rejected the traditional grouping of vertebrates with cephalochordates (P(SH) < 0.01). The recovery of this alternative topology when a shorter branched and more closely related out-group was used suggests that the phylogenetic position of urochordates may be biased by long-branch attraction.
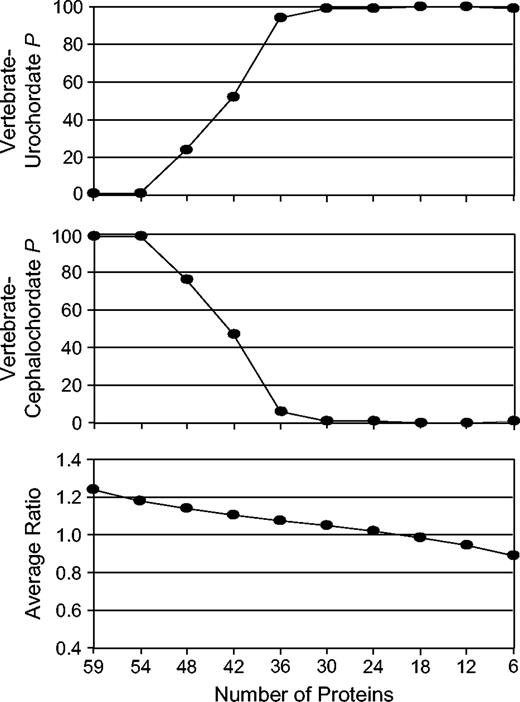
To further test for the possibility of long-branch attraction artifacts, the ratio between the human-urochordate and human-cephalochordate pairwise distances was calculated for each of the 59 proteins used in the initial analysis (fig. 1A). Proteins were then ordered from the highest to the lowest ratio values, and those proteins with higher ratios were sequentially removed in blocks of five to six proteins from the concatenation. Bootstrap support for the resulting NJ topology (Poisson model) was then examined for each data set (fig. 2). As the ratio between the human-urochordate and human-cephalochordate pairwise distances became smaller (closer to one), the topology switched from the traditional grouping of vertebrates and cephalochordates to a well-supported pairing of vertebrates and urochordates. This result suggests that those few proteins with a very high ratio (>1.5) had a large influence on the concatenated topology. As the relative branch lengths became more similar, analytical artifacts such as long-branch attraction may have been overcome to produce the resulting topology.
The effect on bootstrap support for alternative topologies as the ratio between the human-urochordate and human-cephalochordate pairwise distances decreases. Values on the left side of the graphs represent the original concatenation of 59 proteins as presented in fig. 1A. Proteins were then sequentially removed, and bootstrap support (P) for the vertebrate-urochordate (top) and vertebrate-cephalochordate (middle) topologies was evaluated. The average ratio was calculated for the proteins included in each data set (bottom).
In addition, a subset of 15 proteins whose ratio was between 1.1 and 0.9 was concatenated; the resulting topology showed highly significant (>98%) support for urochordates as the closest relative of vertebrates (Supplementary Fig. 1, Supplementary Material online). The likelihood of this topology was also significantly better than the traditional vertebrate + cephalochordate hypothesis (P(SH) < 0.02). These results further suggest that the basal position of urochordates to vertebrates and cephalochordates traditionally found in molecular analyses may be biased by long-branch attraction. A recent study using multiple protein sequences also came to similar conclusions (Philippe, Lartillot, and Brinkmann 2005) and suggested that a vertebrate + urochordate relationship should be considered a plausible hypothesis. Further investigation will be needed to fully resolve the relationship among chordates and may be aided by the completion of the Branchiostoma (cephalochordate) genome.
Phylogenetic analyses of the remaining deuterostome phyla, echinoderms and hemichordates, supported the Ambulacraria hypothesis. With nine proteins, significant (>99%) bootstrap support was found with NJ and ML analyses (Supplementary Fig. 2, Supplementary Material online). Alternative topologies were also significantly rejected with an ML test (P(SH) < 0.01). MP did not recover the grouping of echinoderms with hemichordates but instead showed weak (68%) support for hemichordates as the sister group to vertebrates.
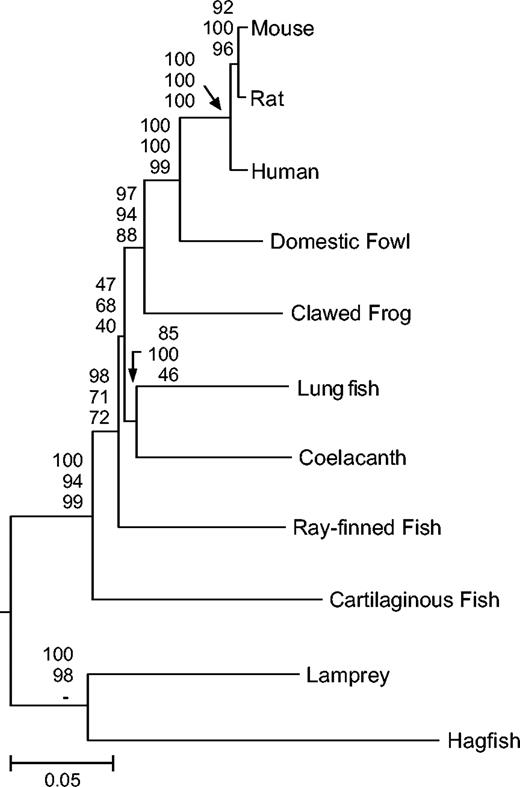
Within vertebrates, most generally accepted relationships were recovered with an analysis of 14 proteins (2,747 amino acids; fig. 3). A close relationship between the lungfish and coelacanth, which together were recovered as the closest relative of tetrapods, received moderate bootstrap support in this phylogeny. Using a larger data set of 41 proteins (9,475 amino acids) with more limited taxonomic sampling, overall bootstrap support for this topology increased (Supplementary Fig. 3, Supplementary Material online), but the log likelihood was not significantly different from an alternative hypothesis placing lungfish alone as the closest relative of tetrapods (−67,718.57 vs. −67,731.98 ± 15, P(SH) = 0.32). Not surprisingly, these results were similar to those of the study responsible for contributing much of the data (Takezaki et al. 2004); this previous study also suggested that the true relationship between tetrapods and the lobe-finned fish may be an unresolvable trichotomy and that an excess of multiple hits may be biasing phylogenetic analyses. Apparently, the divergence in question occurred over a very short time period (approximately 40 Myr, estimated here), creating difficulty in resolving relationships. As suggested previously (Takezaki et al. 2004), larger sequence data sets (>200 proteins) may be required to resolve this relationship, given the rapid divergences between lobe-finned fish and the lineage leading to tetrapods.
Phylogenetic relationships among vertebrates (14 proteins, α = 0.38). NJ topology using a JTT + gamma model is shown. Bootstrap values for NJ (top), ML (middle), and MP (bottom) are shown above each node.
Also within vertebrates, the well-established topology of tetrapods was significantly supported (88%–100% bootstrap support). Cartilaginous fish were recovered as the most basal gnathostomes with moderate bootstrap support (see also Supplementary Fig. 4, Supplementary Material online), although alternative hypotheses could not be rejected with an ML test of 65 proteins (ray-finned fish basal P(SH) = 0.1, Pisces P(SH) = 0.52). Cyclostomata, the pairing of lampreys and hagfish, was significantly supported (see also Supplementary Fig. 5, Supplementary Material online); an alternative hypothesis suggesting lampreys as the sister group to gnathostomes was significantly rejected with a data set of 25 proteins (P(SH) < 0.01).
Divergence Time Analyses
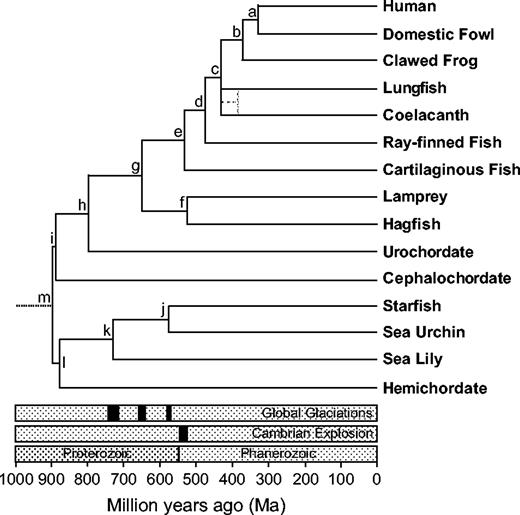
The phylogenetic framework of deuterostome evolution presented above was used to estimate divergence times with a Bayesian local clock method (fig. 4, table 1). Previous studies have estimated deep, Precambrian divergences between deuterostomes and protostomes (Runnegar 1982; Wray, Levinton, and Shapiro 1996; Feng, Cho, and Doolittle 1997; Gu 1998; Wang, Kumar, and Hedges 1999; Nei, Xu, and Glazko 2001; Hedges et al. 2004). Our results suggest that deuterostomes originated prior to approximately 900 MYA and that the first divergence between Ambulacraria and the chordates occurred 896 (832–1,022; 95% credibility interval) MYA. Some studies have produced considerably younger divergence times among deuterostomes (Aris-Brosou and Yang 2002, 2003; Douzery et al. 2004) but suffer from methodological biases, causing their results to conflict with other well-supported aspects of the eukaryote fossil record (Blair and Hedges 2005; Ho et al. 2005).
A Bayesian timescale for deuterostome evolution. Divergence times for each node are presented in table 1. Global glaciations are hypothesized to have occurred approximately 750–700 MYA (Sturtian), 665–635 MYA (Marinoan), and 580 MYA (Varanger/Gaskiers) (Hoffmann et al. 2004). The Cambrian Explosion of fossils occurred approximately 543–520 MYA.
The divergence between echinoderms and hemichordates was estimated at 876 (725–1,074) MYA. The large credibility interval on this divergence was most likely the result of the limited size of the data set (nine proteins; 2,382 amino acids) and may be more tightly constrained as additional sequences becomes available. Within the echinoderms, the divergence between starfish and sea urchins was estimated at 581 (500–669) MYA using 19 proteins (6,813 amino acids); only three sequences were available for estimating the divergence time of sea lilies, so this divergence is presented as the midpoint between the two adjacent nodes (table 1).
Cephalochordates were estimated to have diverged from the other chordates 891 (810–1,067) MYA with an analysis of 75 proteins (23,696 amino acids). Using the complete genome of C. intestinalis (325 proteins; 102,110 amino acids), a divergence time of 794 (685–918) MYA was estimated for the split between vertebrates and urochordates. While the credibility intervals on these two divergences overlap, the increase in age between the vertebrate-urochordate and the vertebrate-cephalochordate splits was consistent with the phylogenetic results presented above. A previous study showing a younger age for the vertebrate-cephalochordate split (750 ± 32 MYA) did not estimate the vertebrate-urochordate divergence in the analysis (Hedges 2001).
Within vertebrates, the divergence between gnathostomes and agnathans was estimated as 652 (605–742) MYA, which was slightly older than previous estimates between 550 and 600 MYA (Wray, Levinton, and Shapiro 1996; Kumar and Hedges 1998). However, our result was based on 51 proteins (17,160 amino acids), which represents 5–10 times more data than previous analyses. The divergence between lampreys and hagfish estimated here at 520 (461–596) MYA was similar to a previous estimate of 499 ± 37 MYA (Hedges 2001), although different data and methods were used (25 proteins; 6,895 amino acids here). Cartilaginous fish were estimated to have diverged from bony fish at 525 (494–580) MYA based on 65 proteins (20,537 amino acids), similar to a previous estimate of 528 ± 56 MYA (Kumar and Hedges 1998). We also estimated that the lineage leading to tetrapods diverged from the other lobe-finned fish 430 (421–438) MYA, while the coelacanth and lungfish diverged 389 (373–404) MYA. However, due to the ambiguity in the phylogenetic analyses, this divergence is shown as an unresolved trichotomy (fig. 4). The divergence between amphibians and amniotes, based on a large data set of 191 proteins (81,117 amino acids), approached the maximum constraint of 370 MYA. When this maximum constraint was removed, the divergence time was estimated at 392 (386–398) MYA.
A total of 325 shared proteins (102,110 amino acids) were recovered from the complete genomes of a limited number of vertebrates and were used to estimate divergence times (see also Supplementary Fig. 6, Supplementary Material online). Our estimate of the divergence between tetrapods and ray-finned fish at 476 (442–494) MYA was slightly older than that of a previous study (450 ± 36 MYA) (Kumar and Hedges 1998), although the confidence intervals of these two estimates overlap. The recent completion of the chicken genome sequence allowed for its use as a minimum fossil constraint (310 MYA), and the divergence time between birds and mammals was estimated at 326 (311–354) MYA.
For comparison, multigene global clock analyses were also performed. On average, divergence times among groups were approximately 20% older under the global clock (Supplementary Table 2, Supplementary Material online). The use of multiple fossil constraints may explain why those times estimated with a Bayesian local clock methods were younger than those estimated with a single (global) evolutionary rate. Divergence time estimates from global clock analyses of small (<20 proteins) data sets are also subject to greater stochasticity, as noted elsewhere (Kumar and Hedges 1998; Hedges et al. 2004).
Our Bayesian timescale showed that all major lineages of deuterostomes had originated before the Cambrian Explosion in the fossil record (∼520 MYA). Previous molecular clock studies have also found deep, Precambrian divergences between animal phyla (Runnegar 1982; Wray, Levinton, and Shapiro 1996; Feng, Cho, and Doolittle 1997; Gu 1998; Wang, Kumar, and Hedges 1999; Nei, Xu, and Glazko 2001; Hedges et al. 2004). The presence of fossil urochordates (Chen et al. 2003), cephalochordates (Shu, Conway Morris, and Zhang 1996), agnathans (Shu et al. 1999), and crinoids (Smith 1988) in Cambrian deposits also argues that the major lineages of deuterostomes had originated prior to the Precambrian-Cambrian boundary. What explains the apparent 300-Myr gap in the deuterostome fossil record? Stem lineages could have originated and persisted for long periods of time before the diversification of modern groups (Hedges 2001). Also, recent geochemical studies have suggested that major shifts in seawater ion concentration, particularly an increase in calcium, occurred approximately 545 MYA (Brennan, Lowenstein, and Horita 2004). The increased availability of calcium may have spurned the evolution of biocalcification in different animal lineages that had originated much earlier.
Some groups, such as hemichordates, cephalochordates, and urochordates, had already diverged prior to global “Snowball Earth” glaciation events in the Neoproterozoic. As many as three major glacial cycles have been suggested between approximately 750 and 545 MYA (see Hoffmann et al. 2004 and references therein) (fig. 4), and their proximity to the Cambrian Explosion has led some to suggest that animal phyla originated as a result of the glaciations (e.g., Hoffman et al. 1998). In contrast, the extent of glaciation into the tropics has been questioned by simulation studies (Hyde et al. 2000; Poulsen 2003) and by other evidence from the geologic record (Condon, Prave, and Benn 2002). If large areas of open water existed during the Neoproterozoic, long periods of glaciation, even as long as 12 Myr (Bodiselitsch et al. 2005), may not have had such a serious impact on global biota. Many eukaryotic lineages certainly must have survived major changes in global climate, and fossil evidence has suggested that complex microbiota may have even flourished in some areas during Snowball Earth times (Corsetti, Awramik, and Pierce 2003).
Conclusions
Here we have addressed some specific phylogenetic questions within deuterostomes and estimated a molecular timescale for the divergences among major lineages. We have shown that the traditional (basal) placement of urochordates observed in previous molecular studies may be an artifact of long-branch attraction. The finding of a close relationship between urochordates and vertebrates suggests that some interpretations of morphological evolution among chordates require reevaluation. Our results also support the pairing of echinoderms with hemichordates as Ambulacraria, corroborating morphological interpretations of larval similarities between these two groups. Within vertebrates, lampreys and hagfish were confirmed to be closely related (Cyclostomata) and the closest relatives of gnathostomes. Our analyses suggest that the coelacanth and lungfish form a group that is the closest living relative of tetrapods and that cartilaginous fish are the most basal gnathostomes. Both of these results, however, did not receive unambiguous support and will require further analysis with larger molecular data sets. Our Bayesian timescale suggests that deuterostomes originated in the Proterozoic and that the major lineages had diverged prior to global glaciation events and the Cambrian Explosion of fossils. By establishing well-supported phylogenies and evolutionary timescales, the interplay between geological events and biological innovations can be examined.
Takashi Gojobori, Associate Editor
We thank David Geiser and two anonymous reviewers for critically reading an earlier version of this manuscript. We also thank Fabia Battistuzzi and Jennifer Sander for assistance with data collection. This work was supported by grants to S.B.H. from the National Science Foundation and the NASA Astrobiology Institute.
References
Adams, M. D., S. E. Celniker, R. A. Holt et al. (195 co-authors).
Ahlberg, P. E., and A. R. Milner.
Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman.
Aparicio, S., J. Chapman, E. Stupka et al. (41 co-authors).
Aris-Brosou, S., and Z. Yang.
———.
Arnason, U., A. Gullberg, and A. Janke.
Arnason, U., A. Gullberg, A. Janke, J. Joss, and C. Elmerot.
Benton, M. J.
Blair, J. E., and S. B. Hedges.
Blair, J. E., P. Shah, and S. B. Hedges.
Bodiselitsch, B., C. Koeberl, S. Master, and W. U. Reimold.
Bourlat, S. J., C. Nielsen, A. E. Lockyer, D. T. Littlewood, and M. J. Telford.
Brennan, S. T., T. K. Lowenstein, and J. Horita.
Brinkmann, H., A. Denk, J. Zitzler, J. J. Joss, and A. Meyer.
Brinkmann, H., B. Venkatesh, S. Brenner, and A. Meyer.
Bromham, L. D., and B. M. Degnan.
Cameron, C. B., J. R. Garey, and B. J. Swalla.
Cao, Y., P. J. Waddell, N. Okada, and M. Hasegawa.
Castresana, J., G. Feldmaier-Fuchs, S. Yokobori, N. Satoh, and S. Paabo.
Chen, J.-Y., D. J. Bottjer, P. Oliveri, S. Q. Dornbos, F. Gao, S. Ruffins, H. Chi, C.-W. Li, and E. H. Davidson.
Chen, J.-Y., D.-Y. Huang, Q.-Q. Peng, H.-M. Chi, X.-Q. Wang, and M. Feng.
Chen, J.-Y., P. Oliveri, C.-W. Li, G.-Q. Zhou, F. Gao, J. W. Hagadorn, K. J. Peterson, and E. H. Davidson.
Clack, J. A.
Condon, D., M. Zhu, S. Bowring, W. Wang, A. Yang, and Y. Jin.
Condon, D. J., A. R. Prave, and D. I. Benn.
Corsetti, F. A., S. M. Awramik, and D. Pierce.
Dehal, P., Y. Satou, R. K. Campbell et al. (87 co-authors).
Delarbre, C., C. Gallut, V. Barriel, P. Janvier, and G. Gachelin.
Donoghue, P. C., P. L. Forey, and R. J. Aldridge.
Donoghue, P. C. J., M. P. Smith, and I. J. Sansom.
Douzery, E. J. P., E. A. Snell, E. Bapteste, F. Delsuc, and H. Philippe.
Feng, D. F., G. Cho, and R. F. Doolittle.
Field, K. G., G. J. Olsen, D. J. Lane, S. J. Giovannoni, M. T. Ghiselin, E. C. Raff, N. R. Pace, and R. A. Raff.
Furlong, R. F., and P. W. H. Holland.
Gibbs, R. A., G. M. Weinstock, M. L. Metzker et al. (228 co-authors).
Giribet, G., D. L. Distel, M. Polz, W. Sterrer, and W. C. Wheeler.
Gu, X.
Halanych, K. M.
Hedges, S. B.
Hedges, S. B., J. E. Blair, M. L. Venturi, and J. L. Shoe.
Hedges, S. B., C. A. Hass, and L. R. Maxson.
Hedges, S. B., and S. Kumar.
Hedges, S. B., P. H. Parker, C. G. Sibley, and S. Kumar.
Hillier, L. W., W. Miller, E. Birney et al. (175 co-authors).
Ho, S. Y., M. J. Phillips, A. J. Drummond, and A. Cooper.
Hoffman, P. F., A. J. Kaufman, G. P. Halverson, and D. P. Schrag.
Hoffmann, K.-H., D. J. Condon, S. A. Bowring, and J. L. Crowley.
Hyde, W. T., T. J. Crowley, S. K. Baum, and W. R. Peltier.
Jacobs, L. L., and W. R. Downs.
Jaillon, O., J.-M. Aury, F. Brunet et al. (61 co-authors).
Janies, D.
Jefferies, R. P. S.
Kishino, H., J. L. Thorne, and W. J. Bruno.
Kumar, S., and S. B. Hedges.
Kuraku, S., D. Hoshiyama, K. Katoh, H. Suga, and T. Miyata.
Kumar, S., K. Tamura, and M. Nei.
Lander, E. S., L. M. Linton, B. Birren et al. (255 co-authors).
Littlewood, D. T., A. B. Smith, K. A. Clough, and R. H. Emson.
Long, J. A., and M. S. Gordon.
Mallatt, J., and J. Sullivan.
Mallatt, J., J. Sullivan, and C. J. Winchell.
Martin, A.
Metschnikoff, V. E.
Meyer, A., and S. I. Dolven.
Meyer, A., and A. C. Wilson.
Nei, M., P. Xu, and G. Glazko.
Nielsen, C.
Northcutt, R. G.
Oda, H., H. Wada, K. Tagawa, Y. Akiyama-Oda, N. Satoh, T. Humphreys, S. Zhang, and S. Tsukita.
Paton, R. L., T. R. Smithson, and J. A. Clack.
Peterson, K. J.
Philippe, H., N. Lartillot, and H. Brinkmann.
Poulsen, C. J.
Rasmussen, A. S., and U. Arnason.
———.
Rasmussen, A. S., A. Janke, and U. Arnason.
Rosen, D. E., P. L. Forey, B. G. Gardiner, and C. Patterson.
Runnegar, B. N.
Ruta, M., and M. I. Coates.
Shimodaira, H., and M. Hasegawa.
Shu, D.-G., S. Conway Morris, and X. L. Zhang.
Shu, D.-G., H.-L. Luo, S. Conway Morrise, X.-L. Zhang, S.-X. Hu, L. Chen, J. Han, M. Zhu, Y. Li, and L.-Z. Chen.
Smith, A. B.
Stock, D. W., K. D. Moberg, L. R. Maxson, and G. S. Whitt.
Stock, D. W., and G. S. Whitt.
Strimmer, K., and A. von Haeseler.
Suga, H., D. Hoshiyama, S. Kuraku, K. Katoh, K. Kubokawa, and T. Miyata.
Suzuki, M., K. Kubokawa, H. Nagasawa, and A. Urano.
Takezaki, N., F. Figueroa, Z. Zaleska-Rutczynska, and J. Klein.
Takezaki, N., F. Figueroa, Z. Zaleska-Rutczynska, N. Takahata, and J. Klein.
Telford, M., and P. Holland.
Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins.
Tohyama, Y., T. Ichimiya, H. Kasama-Yoshida, Y. Cao, M. Hasegawa, H. Kojima, Y. Tamai, and T. Kurihara.
Turbeville, J., J. Schulz, and R. Raff.
Venkatesh, B., M. V. Erdmann, and S. Brenner.
Venter, J. C., M. D. Adams, E. W. Myers et al. (274 co-authors).
Wada, H., and N. Satoh.
Wang, D. Y., S. Kumar, and S. B. Hedges.
Waterston, R. H., K. Lindblad-Toh, E. Birney et al. (222 co-authors).
Winchell, C. J., J. Sullivan, C. B. Cameron, B. J. Swalla, and J. Mallatt.
Winnepenninckx, B., T. Backeljau, and R. De Wachter.
Wray, G. A., J. S. Levinton, and L. H. Shapiro.
Yang, Z.
Yokobori, S., M. Hasegawa, T. Ueda, N. Okada, K. Nishikawa, and K. Watanabe.
Zardoya, R., Y. Cao, M. Hasegawa, and A. Meyer.
Zardoya, R., and A. Meyer.
———.