-
PDF
- Split View
-
Views
-
Cite
Cite
Yoko Fukuda, Masaru Tomita, Takanori Washio, Comparative study of overlapping genes in the genomes of Mycoplasma genitalium and Mycoplasma pneumoniae, Nucleic Acids Research, Volume 27, Issue 8, 1 April 1999, Pages 1847–1853, https://doi.org/10.1093/nar/27.8.1847
Close - Share Icon Share
Abstract
Overlapping genes are defined, in this paper, as a pair of adjacent genes whose coding regions are partly overlapping. We systematically analyzed all overlapping genes in the genomes of two closely related species: Mycoplasma genitalium and Mycoplasma pneumoniae. Careful comparisons were made for homologous genes that are overlapped in one species but not in the other. This comparative analysis allows us to propose a model of how overlapping genes emerged in the course of evolution. It was found that overlapping genes were generated primarily due to the loss of a stop codon in either gene, in many cases, the absence of which resulted in elongation of the 3′ end of the gene's coding region. More specifically, the loss of the stop codon took place as a result of the following events: deletion of the stop codon (64.4%), point mutation at the stop codon (4.4%), and frame shift at the end of the coding region (6.7%). Overlapping genes, in a sense, can be thought of as the results of evolutionary pressure to minimize genome size. However, our analysis indicates that many overlapping genes, at least in the genomes of M.genitalium and M.pneumoniae, are due to incidental elongation of the coding regions.
Introduction
Many overlapping genes have been identified in the genomes of prokaryotes, bacteriophages, animal viruses and mitochondria, some of which have been reported to have functional roles such as in translational coupling (1–5) and negative translational coupling (6,7). Nevertheless, their evolutionary origin, i.e. how they have emerged, is not clearly understood.
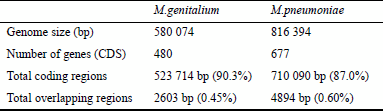
We systematically analyzed all overlapping genes in genomes of two closely related species (Table 1). Mycoplasma genitalium (8) and Mycoplasma pneumoniae (9) were selected for our analysis, as the evolutionary distance of these two species is the closest among the 17 species whose complete genomes are currently available (as of October 1998). Many parts of these two genomes (Fig. 1) are highly homologous; almost all of the genes in M.genitalium are present in M.pneumoniae too, and partial orders of genes are often identical in these two genomes (10).
There are 162 overlapping gene pairs in the genome of M.genitalium according to the TIGR annotation. The genome of M.pneumoniae, on the other hand, contains 203 overlapping gene pairs. There are 135 homologous overlapping gene pairs which exist in both species. The other 27 and 68 overlapping gene pairs are found only in M.genitalium and M.pneumoniae, respectively. The comparative analysis of these two genomes allows us to propose a model of how overlapping genes have emerged over the course of evolution. In particular, careful comparisons were made for the homologous genes that are overlapped in one species but not in the other.
Materials and Methods
The whole genome sequence of M.genitalium with annotation (updated in 1998) was downloaded from the TIGR Microbe Database (http://www.tigr.org/tdb/mdb/mdb.html, updated version), and that of M.pneumoniae was from The Mycoplasma Pneumoniae Genome Project (http://mail.zmbh.uni-heidelberg.de/M-Pneumoniae/MP-Home.html). Information on homologous parts of these two genomes was also obtained from The Mycoplasma Pneumoniae Genome Project.
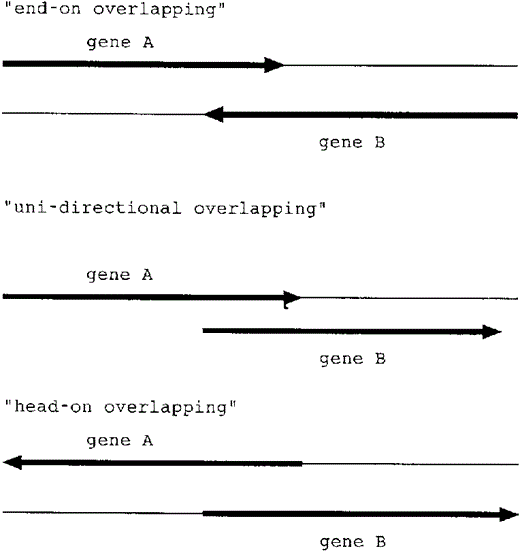
In this paper, overlapping genes are defined as a pair of adjacent genes whose coding regions are partly overlapping. We first list all overlapping genes in their genomes according to the annotations in the databases. For each overlapping gene pair in one species, we aligned the sequence, using ClustalW (11), with the sequence of the homologous part of the other species. We then classified all the cases according to the three directional patterns as described in Figure 1: ‘end-on’, ‘uni-directional’ and ‘head-on’.
For those genes that overlap in one species but not in the other, we made careful analyses in order to infer the cause of the overlapping. Inferred causes of these events of overlapping were then classified into several types.
Further analyses were conducted for those genes whose 3′ ends were elongated by more than 15 amino acids compared with their homologous genes in the other species. FASTA (12) (GenBank version Release 109.0) was used for searching homologous genes in bacteria other than Mycoplasma, and homology of the elongated regions was inspected to see if the regions contain any functionally important sequences. Furthermore, MOTIFS (GCG program package version Unix-8.1 of the Genetics Computer Group, WI) was used for examining possible motifs in these regions.
Annotation of homologous genes is sometimes not in agreement between the two genomes. In particular, many annotational differences were found in determining start codons; i.e. the beginnings of coding regions. This is because the sequence annotations of these two species were made by different software: BLAZE (13) for M.genitalium and FRAMES (GCG program package version Unix-8.1 of the Genetics Computer Group, WI) for M.pneumoniae. We excluded from our discussion those homologous genes whose start codon was assigned differently by the two software packages.
Results and Discussion
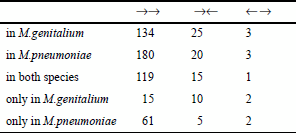
Table 2 summarizes the numbers of overlapping gene pairs in these two genomes. Most overlapping genes are uni-directional, though there are a few ‘end-on’ overlapping genes. Interestingly, there is only one case of a ‘head-on’ overlapping gene.
Out of the ‘end-on’ overlapping genes (the direction of which is →←), many overlap only 1 or 4 bases. Of the 45 overlapping gene pairs (two species together), 16 overlap only 4 bases (Table 3). Mycoplasma genitalium and M.pneumoniae use TAA and TAG for their stop codons. As shown in Figure 2, the complimentary sequence of the stop codon in one gene always includes ‘TA’, which can be a part of the stop codon, TAA or TAG, in the other strand. This explains the large number of 4-base overlapping genes.
Out of the 314 uni-directional overlapping gene pairs (→→), 91 (29.0%) are overlapping only 1 base (Table 4). The overlapped base is either the middle ‘A’ in the sequence ‘TAATG’, which includes TAA for a stop codon of one gene and ATG for a start codon of the other, or the middle ‘G’ in ‘TAGTG’, which includes TAG for the stop codon and GTG for the start codon.
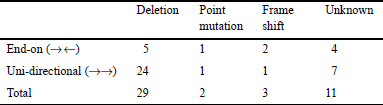
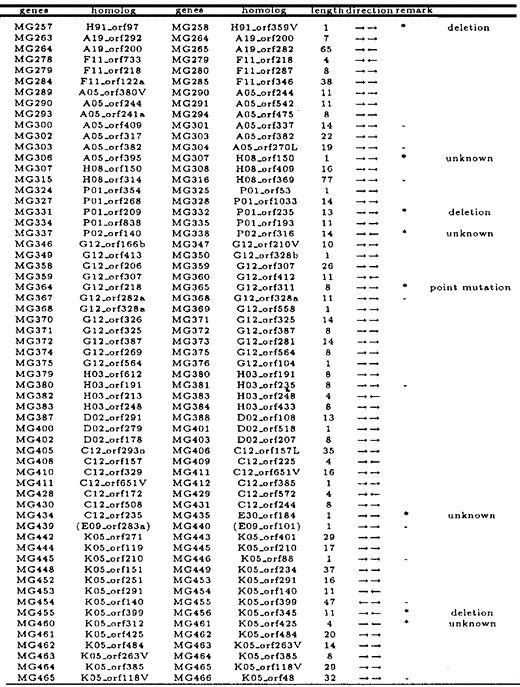
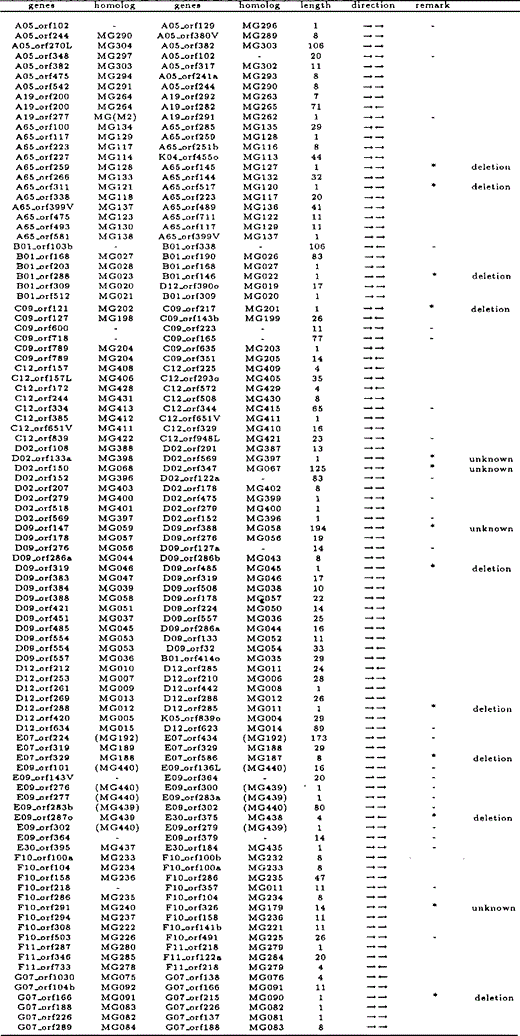
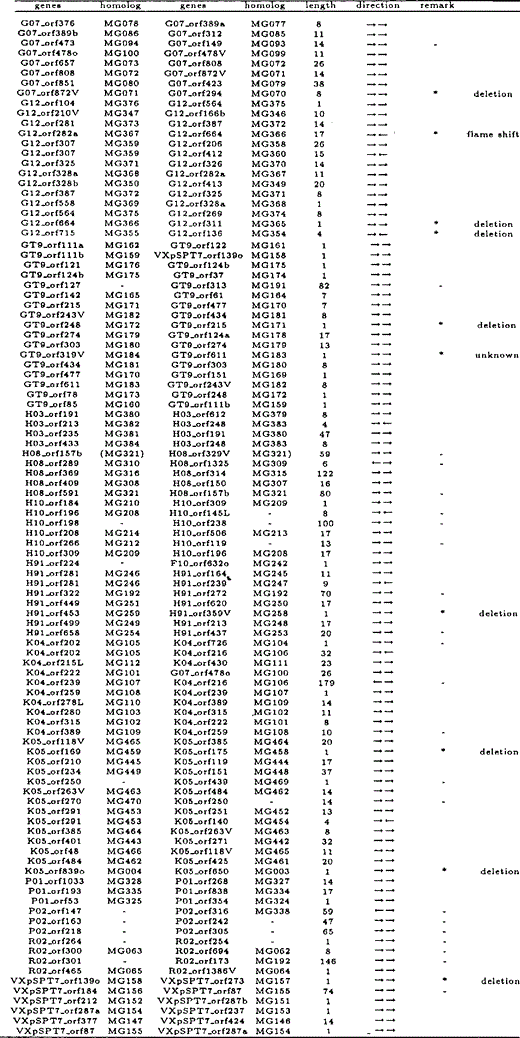
The cause of each case of gene overlapping in one species was inferred from the non-overlapping gene sequence in the other species, and categorized as described with examples in Figure 3a–c. It was found that such overlapping genes were generated primarily due to the loss of a stop codon of either gene, the absence of which resulted in elongation of the 3′ end of the gene's coding region. More specifically, the loss of the stop codon occurred as a result of the following events: deletion of the stop codon (64.4%), point mutation at the stop codon (4.4%), or frame shift at the end of the coding region (6.7%). The results are summarized in Table 5.
Overlapping genes in M.genitalium
Estimated sequence error rate was reported to be <1 in 10 000 bases in M.genitalium (8). The probability of a particular stop codon being replaced due to sequence error is, thus, one in thousands. We therefore consider that sequence errors do not influence the results of our analyses.
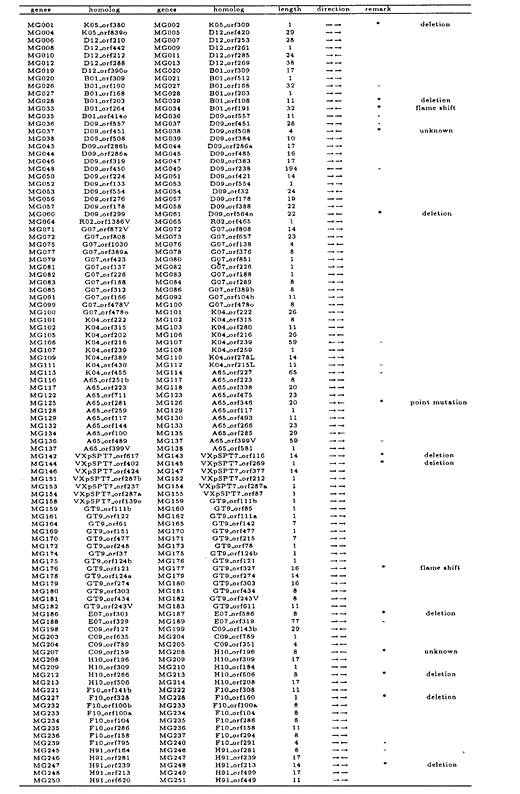
All overlapping gene pairs in M.genitalium and M.pneumoniae, and their homologous genes in the other species, length of overlapping regions, and direction of genes are listed in Tables 6 and 7. Those genes that overlap in one species but not in the other are indicated by an asterisk in the ‘remark’ column. Genes marked with ‘—’ in the column are those which we excluded from our analyses due to annotational difference or absence of homologous genes.
While there is a total of 30 ‘end-on’ overlapping gene pairs (→←), there are only five ‘head-on’ overlapping gene pairs (←→). In addition, most uni-directional overlapping genes (→→) were caused by elongation of the 3′ ends of the preceding genes, not by elongation of the 5′ ends of the subsequent genes. From these observations, we conclude that many overlapping genes were caused by elongation of the 3′ end of a coding region, nearly concomitant with the loss of its stop codon.
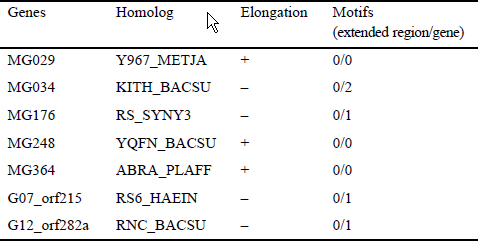
There are seven cases in which gene elongation in one species has presumably occurred by more than 15 amino acids (Table 8). The FASTA search revealed that, for three of the seven cases, certain elongation was also found in other bacteria. However, the elongated regions are not well conserved between the species. Furthermore, the MOTIFS search found no known motifs in the elongated regions for all the seven cases. These results suggest that the elongated regions have little or no functional role that is biologically important.
Overlapping genes might have been thought of as the results of evolutionary pressure to minimize genome size. However, our analysis indicates that many overlapping genes, at least in the genomes of M.genitalium and M.pneumoniae, are due primarily to incidental elongation of coding regions.
Overlapping genes in M.pneumoniae
Causes of overlapping genes. (a) Deletion of stop codon. Deletion of a segment that includes a stop codon in one of two adjacent non-overlapping genes can result in overlapping genes. (b) Point mutation at stop codon. A stop codon (TAA or TAG) in one of two adjacent non-overlapping genes has been lost due to a point mutation, elongating the gene's coding region and resulting in overlapping genes. (c) Frame-shift. Frame-shift mutation in coding region of one of two adjacent non-overlapping genes can cause elongation of the gene, resulting in overlapping genes.
Acknowledgements
We thank Rintaro Saito and Masahiko Wada for their support in computer programing. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas ‘Genome Science’ from The Ministry of Education, Science, Sports and Culture in Japan.



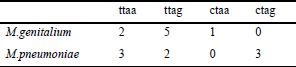
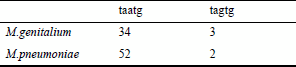


Comments