-
PDF
- Split View
-
Views
-
Cite
Cite
Helen M. Berman, John Westbrook, Zukang Feng, Gary Gilliland, T. N. Bhat, Helge Weissig, Ilya N. Shindyalov, Philip E. Bourne, The Protein Data Bank, Nucleic Acids Research, Volume 28, Issue 1, 1 January 2000, Pages 235–242, https://doi.org/10.1093/nar/28.1.235
Close - Share Icon Share
Abstract
The Protein Data Bank (PDB; http://www.rcsb.org/pdb/ ) is the single worldwide archive of structural data of biological macromolecules. This paper describes the goals of the PDB, the systems in place for data deposition and access, how to obtain further information, and near-term plans for the future development of the resource.
Received September 20, 1999; Revised and Accepted October 17, 1999.
INTRODUCTION
The Protein Data Bank (PDB) was established at Brookhaven National Laboratories (BNL) (1) in 1971 as an archive for biological macromolecular crystal structures. In the beginning the archive held seven structures, and with each year a handful more were deposited. In the 1980s the number of deposited structures began to increase dramatically. This was due to the improved technology for all aspects of the crystallographic process, the addition of structures determined by nuclear magnetic resonance (NMR) methods, and changes in the community views about data sharing. By the early 1990s the majority of journals required a PDB accession code and at least one funding agency (National Institute of General Medical Sciences) adopted the guidelines published by the International Union of Crystallography (IUCr) requiring data deposition for all structures.
The mode of access to PDB data has changed over the years as a result of improved technology, notably the availability of the WWW replacing distribution solely via magnetic media. Further, the need to analyze diverse data sets required the development of modern data management systems.
Initial use of the PDB had been limited to a small group of experts involved in structural research. Today depositors to the PDB have varying expertise in the techniques of X-ray crystal structure determination, NMR, cryoelectron microscopy and theoretical modeling. Users are a very diverse group of researchers in biology, chemistry and computer scientists, educators, and students at all levels. The tremendous influx of data soon to be fueled by the structural genomics initiative, and the increased recognition of the value of the data toward understanding biological function, demand new ways to collect, organize and distribute the data.
In October 1998, the management of the PDB became the responsibility of the Research Collaboratory for Structural Bioinformatics (RCSB). In general terms, the vision of the RCSB is to create a resource based on the most modern technology that facilitates the use and analysis of structural data and thus creates an enabling resource for biological research. Specifically in this paper, we describe the current procedures for data deposition, data processing and data distribution of PDB data by the RCSB. In addition, we address the issues of data uniformity. We conclude with some current developments of the PDB.
DATA ACQUISITION AND PROCESSING
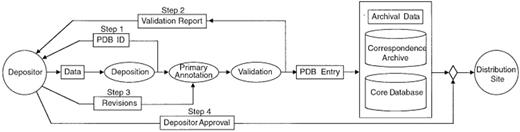
A key component of creating the public archive of information is the efficient capture and curation of the data—data processing. Data processing consists of data deposition, annotation and validation. These steps are part of the fully documented and integrated data processing system shown in Figure 1.
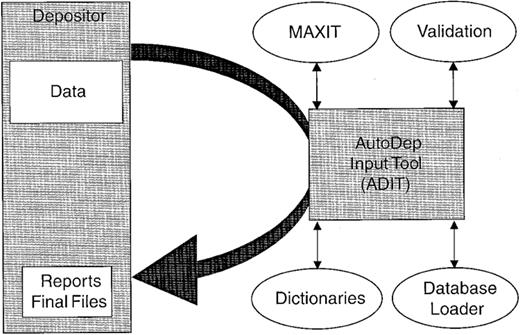
In the present system (Fig. 2), data (atomic coordinates, structure factors and NMR restraints) may be submitted via email or via the AutoDep Input Tool (ADIT; http://pdb.rutgers. edu/adit/ ) developed by the RCSB. ADIT, which is also used to process the entries, is built on top of the mmCIF dictionary which is an ontology of 1700 terms that define the macromolecular structure and the crystallographic experiment (2,3), and a data processing program called MAXIT (MAcromolecular EXchange Input Tool). This integrated system helps to ensure that the data submitted are consistent with the mmCIF dictionary which defines data types, enumerates ranges of allowable values where possible and describes allowable relationships between data values.
After a structure has been deposited using ADIT, a PDB identifier is sent to the author automatically and immediately (Fig. 1, Step 1). This is the first stage in which information about the structure is loaded into the internal core database (see section on the PDB Database Resource). The entry is then annotated as described in the validation section below. This process involves using ADIT to help diagnose errors or inconsistencies in the files. The completely annotated entry as it will appear in the PDB resource, together with the validation information, is sent back to the depositor (Step 2). After reviewing the processed file, the author sends any revisions (Step 3). Depending on the nature of these revisions, Steps 2 and 3 may be repeated. Once approval is received from the author (Step 4), the entry and the tables in the internal core database are ready for distribution. The schema of this core database is a subset of the conceptual schema specified by the mmCIF dictionary.
All aspects of data processing, including communications with the author, are recorded and stored in the correspondence archive. This makes it possible for the PDB staff to retrieve information about any aspect of the deposition process and to closely monitor the efficiency of PDB operations.
Current status information, comprised of a list of authors, title and release category, is stored for each entry in the core database and is made accessible for query via the WWW interface (http://www.rcsb.org/pdb/status.html ). Entries before release are categorized as ‘in processing’ (PROC), ‘in depositor review’ (WAIT), ‘to be held until publication’ (HPUB) or ‘on hold until a depositor-specified date’ (HOLD).
Content of the data collected by the PDB
All the data collected from depositors by the PDB are considered primary data. Primary data contain, in addition to the coordinates, general information required for all deposited structures and information specific to the method of structure determination. Table 1 contains the general information that the PDB collects for all structures as well as the additional information collected for those structures determined by X-ray methods. The additional items listed for the NMR structures are derived from the International Union of Pure and Applied Chemistry recommendations (IUPAC) (4) and will be implemented in the near future.
The information content of data submitted by the depositor is likely to change as new methods for data collection, structure determination and refinement evolve and advance. In addition, the ways in which these data are captured are likely to change as the software for structure determination and refinement produce the necessary data items as part of their output. ADIT, the data input system for the PDB, has been designed so as to easily incorporate these likely changes.
Validation
Validation refers to the procedure for assessing the quality of deposited atomic models (structure validation) and for assessing how well these models fit the experimental data (experimental validation). The PDB validates structures using accepted community standards as part of ADIT’s integrated data processing system. The following checks are run and are summarized in a letter that is communicated directly to the depositor:
Covalent bond distances and angles. Proteins are compared against standard values from Engh and Huber (5); nucleic acid bases are compared against standard values from Clowney et al. (6); sugar and phosphates are compared against standard values from Gelbin et al. (7).
Stereochemical validation. All chiral centers of proteins and nucleic acids are checked for correct stereochemistry.
Atom nomenclature. The nomenclature of all atoms is checked for compliance with IUPAC standards (8) and is adjusted if necessary.
Close contacts. The distances between all atoms within the asymmetric unit of crystal structures and the unique molecule of NMR structures are calculated. For crystal structures, contacts between symmetry-related molecules are checked as well.
Ligand and atom nomenclature. Residue and atom nomenclature is compared against the PDB dictionary (ftp://ftp.rcsb. org/pub/pdb/data/monomers/het_dictionary.txt ) for all ligands as well as standard residues and bases. Unrecognized ligand groups are flagged and any discrepancies in known ligands are listed as extra or missing atoms.
Sequence comparison. The sequence given in the PDB SEQRES records is compared against the sequence derived from the coordinate records. This information is displayed in a table where any differences or missing residues are marked. During structure processing, the sequence database references given by DBREF and SEQADV are checked for accuracy. If no reference is given, a BLAST (9) search is used to find the best match. Any conflict between the PDB SEQRES records and the sequence derived from the coordinate records is resolved by comparison with various sequence databases.
Distant waters. The distances between all water oxygen atoms and all polar atoms (oxygen and nitrogen) of the macromolecules, ligands and solvent in the asymmetric unit are calculated. Distant solvent atoms are repositioned using crystallographic symmetry such that they fall within the solvation sphere of the macromolecule.
In almost all cases, serious errors detected by these checks are corrected through annotation and correspondence with the authors.
It is also possible to run these validation checks against structures before they are deposited. A validation server (http://pdb.rutgers.edu/validate/ ) has been made available for this purpose. In addition to the summary report letter, the server also provides output from PROCHECK (10), NUCheck (Rutgers University, 1998) and SFCHECK (11). A summary atlas page and molecular graphics are also produced.
The PDB will continually review the checking methods used and will integrate new procedures as they are developed by the PDB and members of the scientific community.
Other data deposition centers
The PDB is working with other groups to set up deposition centers. This enables people at other sites to more easily deposit their data via the Internet. Because it is critical that the final archive is kept uniform, the content and format of the final files as well as the methods used to check them must be the same. At present, the European Bioinformatics Institute (EBI) processes data that are submitted to them via AutoDep (http://autodep.ebi.ac.uk/ ). Once these data are processed they are sent to the RCSB in PDB format for inclusion in the central archive. Before this system was put in place it was tested to ensure consistency among entries in the PDB archive. In the future, the data will be exchanged in mmCIF format using a common exchange dictionary, which along with standardized annotation procedures will ensure a high degree of uniformity in the archival data. Structures deposited and processed at the EBI represent ~20% of all data deposited.
Data deposition will also soon be available from an ADIT Web site at The Institute for Protein Research at Osaka University in Japan. At first, structures deposited at this site will be processed by the PDB staff. In time, the staff at Osaka will complete the data processing for these entries and send the files to the PDB for release.
NMR data
The PDB staff recognizes that NMR data needs a special development effort. Historically these data have been retrofitted into a PDB format defined around crystallographic information. As a first step towards improving this situation, the PDB did an extensive assessment of the current NMR holdings and presented their findings to a Task Force consisting of a cross section of NMR researchers. The PDB is working with this group, the BioMagResBank (BMRB) (12), as well as other members of the NMR community, to develop an NMR data dictionary along with deposition and validation tools specific for NMR structures. This dictionary contains among other items descriptions of the solution components, the experimental conditions, enumerated lists of the instruments used, as well as information about structure refinement.
Data processing statistics
Production processing of PDB entries by the RCSB began on January 27, 1999. The median time from deposition to the completion of data processing including author interactions is less than 10 days. The number of structures with a HOLD release status remains at ~22% of all submissions; 28% are held until publication; and 50% are released immediately after processing.
When the RCSB became fully responsible there were about 900 structures that had not been completely processed. These included so called Layer 1 structures that had been processed by computer software but had not been fully annotated. All of these structures have now been processed and are being released after author review.
The breakdown of the types of structures in the PDB is shown in Table 2. As of September 14, 1999, the PDB contained 10 714 publicly accessible structures with another 1169 entries on hold. Of these, 8789 (82%) were determined by X-ray methods, 1692 (16%) were determined by NMR and 233 (2%) were theoretical models. Overall, 35% of the entries have deposited experimental data.
Data uniformity
A key goal of the PDB is to make the archive as consistent and error-free as possible. All current depositions are reviewed carefully by the staff before release. Tables of features are generated from the internal data processing database and checked. Errors found subsequent to release by authors and PDB users are addressed as rapidly as possible. Corrections and updates to entries should be sent to deposit@rcsb. rutgers.edu for the changes to be implemented and re-released into the PDB archive.
One of the most difficult problems that the PDB now faces is that the legacy files are not uniform. Historically, existing data (‘legacy data’) comply with several different PDB formats and variation exists in how the same features are described for different structures within each format. The introduction of the advanced querying capabilities of the PDB makes it critical to accelerate the data uniformity process for these data. We are now at a stage where the query capabilities surpass the quality of the underlying data. The data uniformity project is being approached in two ways. Families of individual structures are being reprocessed using ADIT. The strategy of processing data files as groups of similar structures facilitates the application of biological knowledge by the annotators. In addition, we are examining particular records across all entries in the archive. As an example, we have recently completed examining and correcting the chemical descriptions of all of the ligands in the PDB. These corrections are being entered in the database. The practical consequence of this is that soon it will be possible to accurately find all the structures in the PDB bound to a particular ligand or ligand type. In addition to the efforts of the PDB to remediate the older entries, the EBI has also corrected many of the records in the PDB as part of their ‘clean-up’ project. The task of integrating all of these corrections done at both sites is very large and it is essential that there is a well-defined exchange format to do this; mmCIF will be used for this purpose.
THE PDB DATABASE RESOURCE
The database architecture
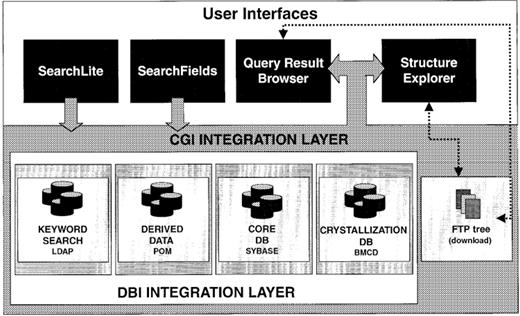
In recognition of the fact that no single architecture can fully express and efficiently make available the information content of the PDB, an integrated system of heterogeneous databases has been created that store and organize the structural data. At present there are five major components (Fig. 3):
• The core relational database managed by Sybase (Sybase SQL server release 11.0, Emeryville, CA) provides the central physical storage for the primary experimental and coordinate data described in Table 1. The core PDB relational database contains all deposited information in a tabular form that can be accessed across any number of structures.
• The final curated data files (in PDB and mmCIF formats) and data dictionaries are the archival data and are present as ASCII files in the ftp archive.
• The POM (Property Object Model)-based databases, which consist of indexed objects containing native (e.g., atomic coordinates) and derived properties (e.g., calculated secondary structure assignments and property profiles). Some properties require no derivation, for example, B factors; others must be derived, for example, exposure of each amino acid residue (13) or Cα contact maps. Properties requiring significant computation time, such as structure neighbors (14), are pre-calculated when the database is incremented to save considerable user access time.
• The Biological Macromolecule Crystallization Database (BMCD; 15) is organized as a relational database within Sybase and contains three general categories of literature derived information: macromolecular, crystal and summary data.
• The Netscape LDAP server is used to index the textual content of the PDB in a structured format and provides support for keyword searches.
It is critical that the intricacies of the underlying physical databases be transparent to the user. In the current implementation, communication among databases has been accomplished using the Common Gateway Interface (CGI). An integrated Web interface dispatches a query to the appropriate database(s), which then execute the query. Each database returns the PDB identifiers that satisfy the query, and the CGI program integrates the results. Complex queries are performed by repeating the process and having the interface program perform the appropriate Boolean operation(s) on the collection of query results. A variety of output options are then available for use with the final list of selected structures.
The CGI approach [and in the future a CORBA (Common Object Request Broker Architecture)-based approach] will permit other databases to be integrated into this system, for example extended data on different protein families. The same approach could also be applied to include NMR data found in the BMRB or data found in other community databases.
Database query
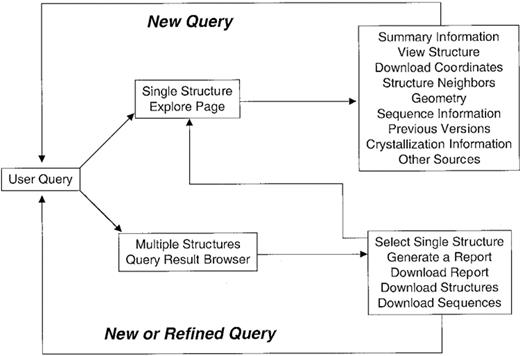
Three distinct query interfaces are available for the query of data within PDB: Status Query (http://www.rcsb.org/pdb/status.html ), SearchLite (http://www.rcsb.org/pdb/searchlite.html ) and SearchFields (http://www.rscb.org/pdb/queryForm.cgi ). Table 3summarizes the current query and analysis capabilities of the PDB. Figure 4 illustrates how the various query options are organized.
SearchLite, which provides a single form field for keyword searches, was introduced in February 1999. All textual information within the PDB files as well as dates and some experimental data are accessible via simple or structured queries. SearchFields, accessible since May 1999, is a customizable query form that allows searching over many different data items including compound, citation authors, sequence (via a FASTA search; 16) and release or deposition dates.
Two user interfaces provide extensive information for result sets from SearchLite or SearchFields queries. The ‘Query Result Browser’ interface allows for access to some general information, more detailed information in tabular format, and the possibility to download whole sets of data files for result sets consisting of multiple PDB entries. The ‘Structure Explorer’ interface provides information about individual structures as well as cross-links to many external resources for macromolecular structure data (Table 4). Both interfaces are accessible to other data resources through the simple CGI application programmer interface (API) described at http://www. rcsb.org/pdb/linking.html
The websiteusage has climbed dramatically since the system was first introduced in February 1999 (Table 5). As of November 1, 1999, the main PDB site receives, on average, greater than one hit per second and greater than one query per minute.
DATA DISTRIBUTION
The PDB distributes coordinate data, structure factor files and NMR constraint files. In addition it provides documentation and derived data. The coordinate data are distributed in PDB and mmCIF formats. Currently, the PDB file is created as the final product of data annotation; the program pdb2cif (17) is used to generate the mmCIF data. This program is used to accommodate the legacy data. In the future, both the mmCIF and PDB format files created during data annotation will be distributed.
Data are distributed to the community in the following ways:
• From primary PDB Web and ftp sites at UCSD, Rutgers and NIST that are updated weekly.
• From complete Web-based mirror sites that contain all databases, data files, documentation and query interfaces updated weekly.
• From ftp-only mirror sites that contain a complete or subset copy of data files, updated at intervals defined by the mirror site. The steps necessary to create an ftp-only mirror site are described in http://www.rcsb.org/pdb/ftpproc.final.html
• Quarterly CD-ROM.
Data are distributed once per week. New data officially become available at 1 a.m. PST each Wednesday. This follows the tradition developed by BNL and has minimized the impact of the transition on existing mirror sites. Since May 1999, two ftp archives have been provided: ftp://ftp.rcsb.org , a reorganized and more logical organization of all PDB data, software, and documentation; and ftp://bnlarchive.rcsb.org , a near-identical copy of the original BNL archive which is maintained for purposes of backward compatibility. RCSB-style PDB mirrors have been established in Japan (Osaka University), Singapore (National University Hospital) and in UK (the Cambridge Crystallographic Data Centre). Plans call for operating mirrors in Brazil, Australia, Canada, Germany, and possibly India.
The first PDB CD-ROM distribution by the RCSB contained the coordinate files, experimental data, software and documentation as found in the PDB on June 30, 1999. Data are currently distributed as compressed files using the compression utility program gzip. Refer to http://www.rcsb.org/pdb/cdrom.html for details of how to order CD-ROM sets. There is presently no charge for this service.
DATA ARCHIVING
The PDB is establishing a central Master Archiving facility. The Master Archive plan is based on five goals: reconstruction of the current archive in case of a major disaster; duplication of the contents of the PDB as it existed on a specific date; preservation of software, derived data, ancillary data and all other computerized and printed information; automatic archiving of all depositions and the PDB production resource; and maintenance of the PDB correspondence archive that documents all aspects of deposition. During the transition period, all physical materials including electronic media and hard copy materials were inventoried and stored, and are being catalogued.
MAINTENANCE OF THE LEGACY BNL SYSTEM
One of the goals of the PDB has been to provide a smooth transition from the system at BNL to the new system. Accordingly, AutoDep, which was developed by BNL (18) for data deposition, has been ported to the RCSB site and enables depositors to complete in-progress depositions as well as to make new depositions. In addition, the EBI accepts data using AutoDep. Similarly, the programs developed at BNL for data query and distribution (PDBLite, 3DBbrowser, etc.) are being maintained by the remaining BNL-style mirrors. The RCSB provides data in a form usable by these mirrors. Finally the style and format of the BNL ftp archive is being maintained at ftp://bnlarchive. rcsb.org
A multitude of resources and programs depend upon their links to the PDB. To eliminate the risk of interruption to these services, links to the PDB at BNL were automatically redirected to the RCSB after BNL closed operations on June 30, 1999 using a network redirect implemented jointly by RCSB and BNL staff. While this redirect will be maintained, external resources linking to the PDB are advised to change any URLs from http://www.pdb.bnl.gov/ to http://www.rcsb.org/
CURRENT DEVELOPMENTS
In the coming months, the PDB plans to continue to improve and develop all aspects of data processing. Deposition will be made easier, and annotation will be more automated. In addition, software for data deposition and validation will be made available for in-laboratory use.
The PDB will also continue to develop ways of exchanging information between databases. The PDB is leading the Object Management Group Life Sciences Initiative’s efforts to define a CORBA interface definition for the representation of macromolecular structure data. This is a standard developed under a strict procedure to ensure maximum input by members of various academic and industrial research communities. At this stage, proposals for the interface definition, including a working prototype that uses the standard, are being accepted. For further details refer to http://www.omg.org/cgi-bin/doc?lifesci/ 99-08-15 . The finalized standard interface will facilitate the query and exchange of structural information not just at the level of complete structures, but at finer levels of detail. The standard being proposed by the PDB will conform closely to the mmCIF standard. It is recognized that other forms of data representation are desirable, for example using eXtensible Markup Language (XML). The PDB will continue to work with mmCIF as the underlying standard from which CORBA and XML representations can be generated as dictated by the needs of the community.
The PDB will also develop the means and methods of communications with the broad PDB user community via the Web. To date we have developed prototype protein documentaries (19) that explore this new medium in describing structure–function relationships in proteins. It is also possible to develop educational materials that will run using a recent Web browser (20).
Finally it is recognized that structures exist both in the public and private domains. To this end we are planning on providing a subset of database tools for local use. Users will be able to load both public and proprietary data and use the same search and exploratory tools used at PDB resources.
The PDB does not exist in isolation, rather each structure represents a point in a spectrum of information that runs from the recognition of an open reading frame to a fully understood role of the single or multiple biological functions of that molecule. The available information that exists on this spectrum changes over time. Recognizing this, the PDB has developed a scheme for the dynamic update of a variety of links on each structure to whatever else can be automatically located on the Internet. This information is itself stored in a database and can be queried. This feature will appear in the coming months to supplement the existing list of static links to a small number of the more well known related Internet resources.
PDB ADVISORY BOARDS
The PDB has several advisory boards. Each member institution of the RCSB has its own local PDB Advisory Committee. Each institution is responsible for implementing the recommendations of those committees, as well as the recommendations of an International Advisory Board. Initially, the RCSB presented a report to the Advisory Board previously convened by BNL. At their recommendation, a new Board has been approached which contains previous members and new members. The goal was to have the Board accurately reflect the depositor and user communities and thus include experts from many disciplines.
Serious issues of policy are referred to the major scientific societies, notably the IUCr. The goal is to make decisions based on input from a broad international community of experts. The IUCr maintains the mmCIF dictionary as the data standard upon which the PDB is built.
FOR FURTHER INFORMATION
The PDB seeks to keep the community informed of new developments via weekly news updates to the Web site, quarterly newsletters, and a soon to be initiated annual report. Users can request information at any time by sending mail to info@rcsb. org . Finally, the pdb-l@rcsb.org listserver provides a community forum for the discussion of PDB-related issues. Changes to PDB operations that may affect the community, for example, data format changes, are posted here and users have 60 days to discuss the issue before changes are made according to major consensus. Table 6indicates how to access these resources.
CONCLUSION
These are exciting and challenging times to be responsible for the collection, curation and distribution of macromolecular structure data. Since the RCSB assumed responsibility for data deposition in February 1999, the number of depositions has averaged approximately 50 per week. However, with the advent of a number of structure genomics initiatives worldwide this number is likely to increase. We estimate that the PDB, which at this writing contains approximately 10 500 structures, could triple or quadruple in size over the next 5 years. This presents a challenge to timely distribution while maintaining high quality. The PDB’s approach of using modern data management practices should permit us to scale to accommodate a large data influx.
The maintenance and further development of the PDB are community efforts. The willingness of others to share ideas, software and data provides a depth to the resource not obtainable otherwise. Some of these efforts are acknowledged below. New input is constantly being sought and the PDB invites you to make comments at any time by sending electronic mail to info@rcsb.org
ACKNOWLEDGEMENTS
Research Collaboratory for Structural Bioinformatics (RCSB) is a consortium consisting of three institutions: Rutgers University, San Diego Supercomputer Center at University of California, San Diego, and the National Institute of Standards and Technology. The current RCSB PDB staff include the authors indicated and Kyle Burkhardt, Anke Gelbin, Michael Huang, Shri Jain, Rachel Kramer, Nate Macapagal, Victoria Colflesh, Bohdan Schneider, Kata Schneider, Christine Zardecki (Rutgers); Phoebe Fagan, Diane Hancock, Narmada Thanki, Michael Tung, Greg Vasquez (NIST); Peter Arzberger, John Badger, Douglas S. Greer, Michael Gribskov, John Kowalski, Glen Otero, Shawn Strande, Lynn F. Ten Eyck, Kenneth Yoshimoto (UCSD). The continuing support of Ken Breslauer (Rutgers), John Rumble (NIST) and Sid Karin (SDSC) is gratefully acknowledged. Current collaborators contributing to the future development of the PDB are the BioMagResBank, the Cambridge Crystallographic Data Centre, the HIV Protease Database Group, The Institute for Protein Research, Osaka University, National Center for Biotechnology Information, the ReLiBase developers, and the Swiss Institute for Bio-informatics/Glaxo. We are especially grateful to Kim Henrick of the EBI and Steve Bryant at NCBI who have reviewed our files and sent back constructive criticisms. This has helped the PDB to continuously improve its procedures for producing entries. The cooperation of the BNL PDB staff is gratefully acknowledged. Portions of this article will appear in Volume F of the International Tables of Crystallography. This work is supported by grants from the National Science Foundation, the Office of Biology and Environmental Research at the Department of Energy, and two units of the National Institutes of Health: the National Institute of General Medical Sciences and the National Institute of Medicine.
To whom correspondence should be addressed at: Department of Chemistry, Rutgers University, 610 Taylor Road, Piscataway, NJ 08854-8087, USA. Tel: +1 732 445 4667; Fax: +1 732 445 4320; Email: berman@rcsb.rutgers.edu
Figure 1. The steps in PDB data processing. Ellipses represent actions and rectangles define content.
Figure 2. The integrated tools of the PDB data processing system.
Figure 3. The integrated query interface to the PDB.
Figure 4. The various query options that are available for the PDB.
Current query capabilities of the PDB
| TABLE CRC: gkd090to3 |
| TABLE CRC: gkd090to3 |
Current query capabilities of the PDB
| TABLE CRC: gkd090to3 |
| TABLE CRC: gkd090to3 |
Static cross-links to other data resources currently provided by the PDB
| TABLE CRC: gkd090t04 |
| TABLE CRC: gkd090t04 |
Static cross-links to other data resources currently provided by the PDB
| TABLE CRC: gkd090t04 |
| TABLE CRC: gkd090t04 |
Web query statistics for the primary RCSB site (http://www.rcsb.org )
| TABLE CRC: gkd090to5 |
| TABLE CRC: gkd090to5 |
Web query statistics for the primary RCSB site (http://www.rcsb.org )
| TABLE CRC: gkd090to5 |
| TABLE CRC: gkd090to5 |
References
1 Bernstein,F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E., Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (
2 Bourne,P., Berman,H.M., Watenpaugh,K., Westbrook,J.D. and Fitzgerald,P.M.D. (
4 Markley,J.L., Bax,A., Arata,Y., Hilbers,C.W., Kaptein,R., Sykes,B.D., Wright,P.E. and Wüthrich,K. (
6 Clowney,L., Jain,S.C., Srinivasan,A.R., Westbrook,J., Olson,W.K. and Berman,H.M. (
7 Gelbin,A., Schneider,B., Clowney,L., Hsieh,S.-H., Olson,W.K. and Berman,H.M. (
9 Zhang.J., Cousens,L.S., Barr,P.J. and Sprang,S.R. (
10 Laskowski,R.A., McArthur,M.W., Moss,D.S. and Thornton,J.M. (
21 Siddiqui,A. and Barton,G. (
22 Orengo,C.A., Michie,A.D., Jones,S., Jones,D.T., Swindels,M.B. and Thornton,J.M. (
29 Berman,H.M., Olson,W.K., Beveridge,D.L., Westbrook,J., Gelbin,A., Demeny,T., Hsieh,S.H., Srinivasan,A.R. and Schneider,B. (
31 Laskowski,R.A., Hutchinson,E.G., Michie,A.D., Wallace,A.C., Jones,M.L. and Thornton,J.M. (
33 Neshich,G., Togawa,R., Vilella,W. and Honig,B. (




Comments