-
PDF
- Split View
-
Views
-
Cite
Cite
Todd Blevins, Rajendran Rajeswaran, Padubidri V. Shivaprasad, Daria Beknazariants, Azeddine Si-Ammour, Hyun-Sook Park, Franck Vazquez, Dominique Robertson, Frederick Meins, Thomas Hohn, Mikhail M. Pooggin, Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing, Nucleic Acids Research, Volume 34, Issue 21, 1 November 2006, Pages 6233–6246, https://doi.org/10.1093/nar/gkl886
Close - Share Icon Share
ABSTRACT
Like other eukaryotes, plants use DICER-LIKE (DCL) proteins as the central enzymes of RNA silencing, which regulates gene expression and mediates defense against viruses. But why do plants like Arabidopsis express four DCLs, a diversity unmatched by other kingdoms? Here we show that two nuclear DNA viruses (geminivirus CaLCuV and pararetrovirus CaMV) and a cytoplasmic RNA tobamovirus ORMV are differentially targeted by subsets of DCLs. DNA virus-derived small interfering RNAs (siRNAs) of specific size classes (21, 22 and 24 nt) are produced by all four DCLs, including DCL1, known to process microRNA precursors. Specifically, DCL1 generates 21 nt siRNAs from the CaMV leader region. In contrast, RNA virus infection is mainly affected by DCL4. While the four DCLs are partially redundant for CaLCuV-induced mRNA degradation, DCL4 in conjunction with RDR6 and HEN1 specifically facilitates extensive virus-induced silencing in new growth. Additionally, we show that CaMV infection impairs processing of endogenous RDR6-derived double-stranded RNA, while ORMV prevents HEN1-mediated methylation of small RNA duplexes, suggesting two novel viral strategies of silencing suppression. Our work highlights the complexity of virus interaction with host silencing pathways and suggests that DCL multiplicity helps mediate plant responses to diverse viral infections.
INTRODUCTION
RNA silencing in multicellular plants and animals is mediated by ∼21–24 nt small RNAs (sRNAs) that guide sequence-specific gene regulation, chromatin modification, and defense against viruses. These sRNAs are broadly classified into microRNAs (miRNAs) and small interfering RNAs (siRNAs), which have similar chemical structures but differ in function and mode of biogenesis. Production of both types of sRNAs depends on the activity of dicer proteins. Plants such as Arabidopsis have evolved a diversity of RNA silencing pathways, sRNA classes and DICER-LIKE (DCL) genes that is unmatched in other eukaryotes (1–4).
Like their animal counterparts, plant miRNAs are excised from stem-loop structures of primary miRNA gene transcripts by DCL1 in cooperation with its binding partner HYL1 (5,6). The resulting miRNA/miRNA∗ duplex is loaded onto the argonaute family protein AGO1, the slicer component of the RNA-induced silencing complex (RISC) (7), which in turn is guided to the complementary target mRNAs by the miRNA strand.
In contrast to miRNAs, endogenous siRNAs are cleaved from long perfect double-stranded (ds) RNAs, which are themselves products of specific RNA-dependent RNA polymerase (RDR) activities (8). DCL4 and DCL3 process such dsRNA substrates to produce the predominant siRNA size classes, 21 and 24 nt, respectively (3,4). Like miRNAs, certain DCL4-dependent endogenous 21 nt siRNAs silence their target genes in trans (9). These trans-acting siRNAs (ta-siRNAs) associate with AGO-RISC (7) and target complementary mRNAs for cleavage and degradation. The biogenesis of ta-siRNAs depends on miRNA-directed cleavage of non-coding TAS transcripts to generate products that are converted by RDR6 into dsRNA substrates for DCL4 (10–13). The DCL3-dependent 24 nt siRNAs, mostly derived from repetitive DNA loci (repeat-associated siRNAs; ra-siRNA), likely mediate the establishment and maintenance of chromatin states through RNA-dependent DNA methylation and histone modification (2). The biogenesis of ra-siRNAs requires the RNA polymerases POL IVa and RDR2 (14,15). Another argonaute protein, AGO4, is involved in the ra-siRNA pathway, probably as the effector component of the putative RNA-induced transcriptional silencing complex (16,17).
A subsequent step in the biogenesis of all plant sRNAs is methylation of their 3′-terminal nucleotide at the 2′-hydroxyl group by the methyltransferase HEN1 (18–20), which protects them from degradation and oligouridylation (21).
RNA silencing that affects transgene expression can spread locally cell-to-cell and systemically via vascular tissues throughout the plant (22,23). Although the nature of the sequence-specific silencing signal is unknown, cell-to-cell spread of transgene silencing is correlated with DCL4-generated 21 nt siRNAs (24). Interestingly, longer-range cell-to-cell movement of transgene silencing requires RDR6 (25), which is normally coupled to DCL4 for the biogenesis of 21 nt ta-siRNAs. It is speculated that RDR6 is involved in a relay amplification of the silencing signal (25). RDR6 was also implicated in systemic transgene silencing (26). The spread of the silencing signal is a potential component of plant defense against ongoing virus infections, especially for protection of the shoot apical meristem (26).
More generally, RNA silencing may represent an adaptive immune response of plants against viruses (27). sRNAs of distinct size-classes have been detected in plants infected with RNA viruses of different families (28–31) and DNA geminiviruses (19,32). RNA viruses are mainly targeted by DCL4 and DCL2, which produce 21 and 22 nt viral siRNAs (29–31). Our previous work implicated DCL2 and DCL3 in production of 22 and 24 nt geminiviral siRNAs, respectively, and an additional DCL activity producing 21 nt geminiviral siRNAs (19). Neither the function of these geminivirus-derived siRNAs, nor siRNA biogenesis for other DNA viruses has been examined using the genetic resources acquired from the study of endogenous sRNA species.
Viruses modified to carry a sequence homologous to a host-encoded gene are informative tools for studying siRNA-dependent silencing. Such constructs can trigger silencing of the homologous gene at both post-transcriptional and transcriptional levels (27). This phenomenon is called virus induced gene silencing (VIGS). RDR6 and its cofactor SGS3 are important for VIGS triggered by both RNA virus (RNA-VIGS) (27) and DNA geminivirus vectors (DNA-VIGS) (33). Recently, DCL4- and DCL2-dependent viral siRNAs have been implicated in RNA-VIGS (30). Similar to transgene-induced silencing, VIGS can spread cell-to-cell and systemically (27). Peele et al. (34) revealed virus-independent spreading of DNA-VIGS from geminivirus-infected tissues to the shoot apical meristem, from which viruses are normally excluded. The pathways mediating DNA-VIGS have not been thoroughly dissected; given the cytoplasmic and nuclear steps in replication of DNA viruses, they are ideal for studying virus-induced RNA silencing in its entirety. Furthermore, DNA viruses do not code for their own RDRs and are therefore useful for examining the role of host RDRs in different silencing processes.
Despite silencing-based responses to viral infection, plant viruses can still establish robust infection in susceptible hosts, in part by suppressing RNA silencing. Plant viruses encode various suppressor proteins that do not share common features, although many of them bind short or long dsRNA (35,36). For example, the RNA tombusvirus-encoded suppressor P19 binds siRNA duplexes selectively, which might sequester them from the silencing process (37). Viral silencing suppressors can cause defects in endogenous silencing pathways when expressed from transgenes (38), possibly due to their dsRNA binding property.
Here we describe genetic requirements for the biogenesis of 21, 22 and 24 nt siRNAs associated with DNA viruses of the geminivirus [Cabbage leaf curl virus (CaLCuV)] and the pararetrovirus [Cauliflower mosaic virus (CaMV)] families, which form circular minichromosomes in the nucleus (39,40). We contrast this to the predominantly 21 nt siRNAs derived from a cytoplasmic RNA tobamovirus [Oilseed rape mosaic virus (ORMV)] (41). We show that the three viruses are differentially targeted by the four Arabidopsis DCLs producing specific size classes of viral siRNAs. Using a geminivirus VIGS vector (42) and Arabidopsis silencing mutants we find the four DCLs to be partially redundant for DNA-VIGS targeting an endogenous mRNA. However, extensive VIGS in newly emerging tissues requires DCL4 in conjunction with RDR6 and HEN1. As a derivative of this study with CaMV and ORMV, we also uncover two novel viral strategies of silencing suppression.
MATERIALS AND METHODS
Plants and viruses
The following previously described Arabidopsis mutant lines were used: dcl1-8 [in Col-gl1, originally: sin1-2; (43)], dcl1-9 [in La-er; originally: caf-1; (43)], dcl2-5 [in Col-0; (19)], dcl3-1 [in Col-0; (29)], dcl4-2 [in Col-0; (12)], rdr2-1 [in Col-0; (29)], rdr6-15 [in Col-0; (10)], nrpd1a-3 [in Col-0; (14)], ago4-1 [in La-er; (16)], hyl1-2 and hen1-5 [both in Col-0; (44)]. To obtain the double mutant dcl2 dcl3 (d2d3), dcl2-5 was crossed with dcl3-1. Homozygous d2d3 was then crossed with dcl4-2 to obtain homozygous double mutants dcl2 dcl4 (d2d4) and dcl3 dcl4 (d3d4) and the triple mutant dcl2 dcl3 dcl4 (d2d3d4) in the F2 segregating population. Homozygous lines were selected using PCR with allele-specific primers (see Supplementary Data).
Arabidopsis wt and mutant plants were grown from seeds in soil in a phytochamber (Sanyo) at 20°C with 12 h day and 12 h night. Four to five weeks post-germination, unless otherwise stated, seedlings were inoculated with CaLCuV or CaMV by biolistic delivery of a plasmid mixture (0.5 μg each) of pMTCbLCVA.008 and pCPCbLCVB.002 (42) or 1 μg plasmid pCa122 [the CaMV strain CM1841; (45)], respectively, as described earlier (19).
One month post-inoculation, unless otherwise stated, virus-infected plants were harvested in pools and ground in liquid nitrogen for total RNA (19) and DNA (46) preparations. Titers of the viruses were determined by semi-quantitative duplex PCR (see Supplementary Data).
Mechanical inoculation of Arabidopsis plants with ORMV (41) was performed at around 5 weeks post-germination using celite 545 (Merck) and sap of ORMV-infected Nicotiana benthamiana.
RNA blot hybridization
Total RNA preparation, size-fractionation, β-elimination and RNA blot hybridization were performed as described previously (19) (for probes and other details, see Supplementary Data).
Real-time RT–PCR quantification
See Supplementary Data for details of cDNA synthesis, PCR conditions and primers. Uncertainties were propagated from standard errors for triplicate measurements of cDNA pools (derived from the column-purified RNA of 3–4 plant replicates).
RESULTS
Three size classes of DNA virus-derived siRNAs
Using a reverse genetics approach we investigated the RNA silencing-based response of plants to DNA virus infection. In experiments described separately below, we inoculated Arabidopsis plants with the pararetrovirus CaMV (45) and with a recombinant geminivirus CaLCuV, which carries a segment of the Arabidopsis Chlorata I (ChlI) gene (CaLCuV::Chl) (42). The latter construct also allowed us to study the genetic requirements for VIGS.
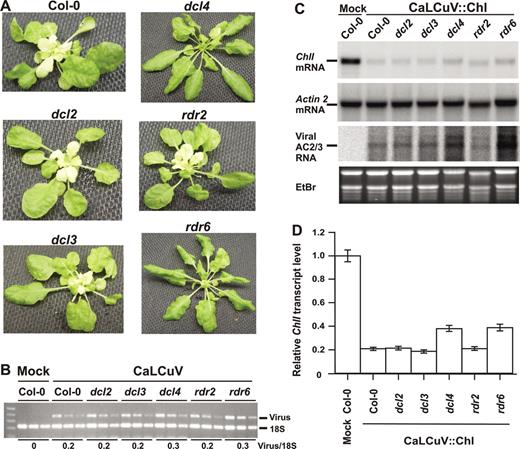
Both viruses established systemic infections in Arabidopsis, accompanied by deformations on young and old leaves one month post-inoculation (Figure 1A and Supplementary Figure S1). As expected, CaLCuV::Chl triggered VIGS of the endogenous ChlI gene, resulting in a yellow-white (‘chlorata’) phenotype in emerging inner rosette leaves (Figure 1A), described earlier by Muangsan et al. (42). Infection with CaLCuV::Chl (Figure 1C), but not with CaMV (data not shown), greatly reduced the ChlI mRNA content of total RNA prepared from whole plants, indicating efficient, sequence-specific VIGS at the whole plant level. Mature leaves that developed before inoculation with CaLCuV::Chl did not exhibit the chlorata phenotype, likely due to residual CHLI protein or its dispensability later in development for maintaining high chlorophyll content.
Mutations in DCL4 and RDR6 impair Chl-VIGS in infected tissue and abolish total Chl-VIGS in newly emerging leaves. (A) wt (Col-0), dcl and rdr mutant plants 1 month post-inoculation with CaLCuV::Chl. Infection causes leaf deformations and a yellow-white phenotype in Col-0 due to Chl-VIGS. Total DNA and RNA were isolated from pooled plants. Viral titers (B) were measured by semi-quantitative PCR on serial dilutions (5-fold each) of the DNA. 18S ribosomal DNA amplification is an internal control. (C) RNA blot hybridization was performed with column-purified total RNA (8 μg/lane). The membrane was successively hybridized with random-labeled DNA probes for ChlI, ACT2 and CaLCuV AC2/AC3 transcripts. (D) Quantitative real-time PCR was made on cDNA synthesized from RNA pools. The mean from triplicate determinations of ChlI transcript levels was normalized to each corresponding ACT2 mean; the Col-0 (mock) level was set to 1.
Wild-type (wt) accessions Col-0, Col-gl1 and La-er infected with CaLCuV::Chl and CaMV accumulated similar patterns of 21, 22 and 24 nt sRNAs of both sense and antisense polarities, from both coding and non-coding regions (Figures 2 and 3). Based on these and other findings for geminiviruses (19), we conclude that both families of DNA viruses produce three distinct size-classes of siRNAs.
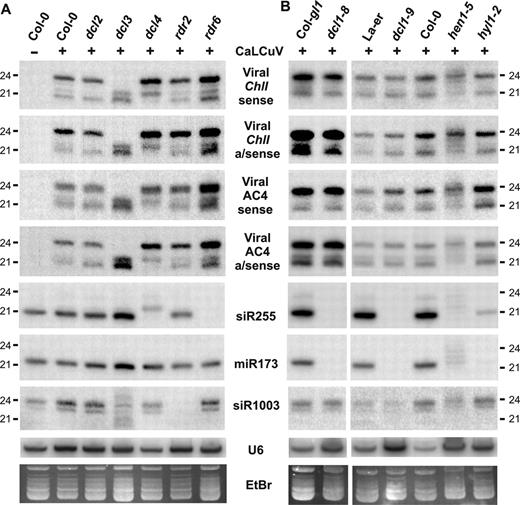
CaLCuV-derived siRNA accumulation in Arabidopsis mutants for RNA silencing pathways. Low molecular weight (LMW) RNA (10 μg/lane) from plant pools was analyzed by RNA blot hybridization. Membranes were successively hybridized with sense and antisense DNA oligo probes for the viral ChlI-fragment, viral AC4 region and endogenous controls (see Supplementary Table S1 for probe details). Synthetic 21 and 24 nt RNA oligos were used as markers. (A) Analysis of mock Col-0 and virus-infected Col-0, dcl- and rdr-mutant plants. (B) Analysis of mock Col-0, virus-infected controls (Col-gl1, La-er, Col-0), and mutants affecting miRNA accumulation (dcl1-8, dcl1-9, hen1-5, hyl1-2). U6 signal and ethidium bromide (EtBr) staining serve as loading controls.
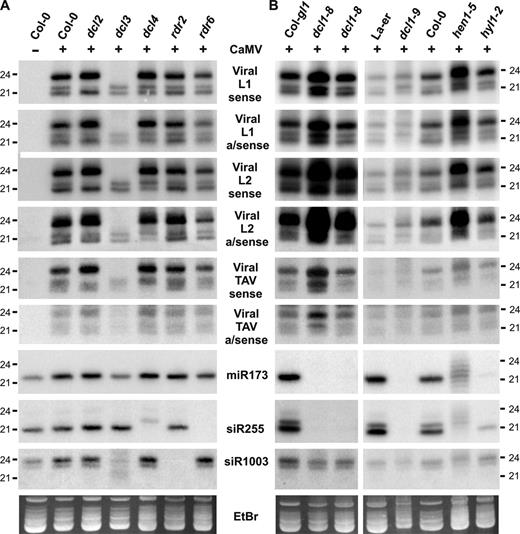
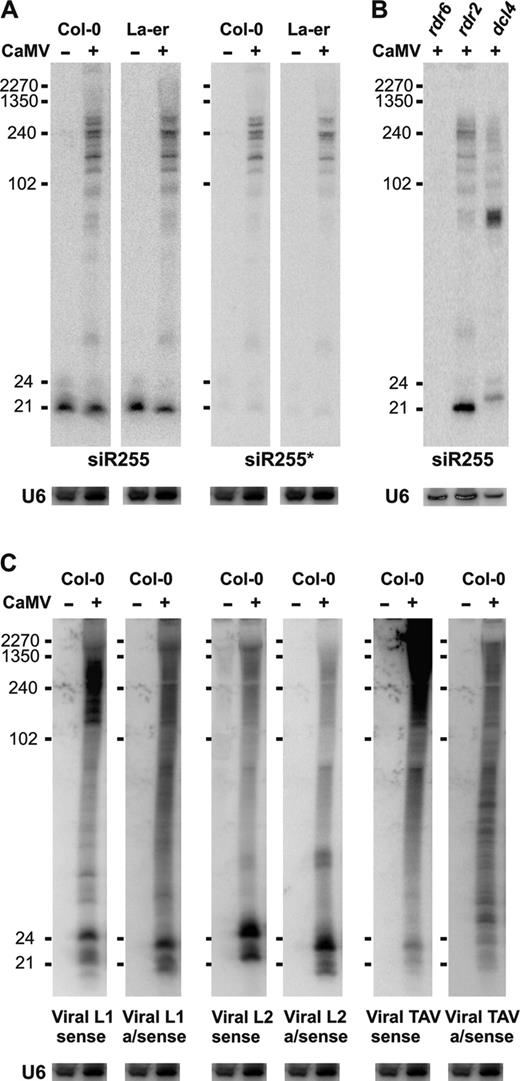
CaMV-derived siRNA accumulation in Arabidopsis mutants for RNA silencing pathways. Small RNA blot analysis was performed as in Figure 2. Membranes were successively hybridized with sense and antisense DNA oligo probes for viral L1 and L2 regions of the CaMV transcript leader, TAV coding region and endogenous controls (see Supplementary Table S1 for probe details). (A) Analysis of Col-0 (mock) and virus-infected Col-0, dcl and rdr-mutant plants. (B) Analysis of mock Col-0, virus-infected controls (Col-gl1, La-er, Col-0), and mutants affecting miRNA accumulation (dcl1-8, dcl1-9, hen1-5, hyl1-2). EtBr staining is a loading control.
Since the levels of siRNAs derived from the CaLCuV-based ChlI sequence and the virus genes (AC4 and AC2) were equally high [Figure 2 and (19)], we assume that most ChlI siRNAs are produced from viral AV1-ChlI chimeric transcripts rather than endogenous ChlI mRNA, which accumulates to lower levels. Further supporting this assertion, secondary siRNAs from ChlI mRNA regions flanking the target sequence were barely detectable (data not shown).
Specific deficiencies in endogenous RNA silencing pathways affect viral siRNA production and impair VIGS
The presence of different size classes of viral siRNAs suggested that multiple RNA silencing pathways respond to DNA viruses. To determine the relative contribution of these pathways, we examined the effect of well-characterized, T-DNA insertion deficiency mutants on the infection process, VIGS, and the accumulation of viral siRNAs. We first focused on mutants of DCL2, DCL3, DCL4, RDR2 and RDR6, which mediate the biogenesis of endogenous siRNAs (4).
None of these mutants differed markedly from wt in development or severity of CaLCuV symptoms (Figure 1A). One month post-inoculation, viral DNA was measured in pools of 3–4 plants by semi-quantitative duplex PCR. CaLCuV DNA accumulation in dcl2, dcl3, and rdr2 was comparable to that of wt, while slightly higher in dcl4 and rdr6 (Figure 1B). Similarly, CaLCuV transcripts measured by RNA blot hybridization were comparable in dcl2, dcl3, rdr2 and wt, and elevated in dcl4 and rdr6 (Figure 1C, viral AC2/3 RNA). Strikingly, dcl4 and rdr6, exhibited almost no chlorata phenotype in new growth, while dcl2, dcl3 and rdr2 developed yellow-white inner rosette leaves similar to the wt infection (Figure 1A). Thus, although none of these mutants had dramatic effects on virus infection, DCL4 and RDR6 were required for extensive Chl-VIGS in newly emerging leaves. Interestingly, DCL4 is required for cell-to-cell spread of transgene-induced silencing in Arabidopsis (24), raising a possibility that this dicer is involved in production of a virus-induced silencing signal as well (see Discussion).
We compared the effect of these mutants on the accumulation of viral siRNAs and ChlI mRNA. The decreased accumulation of siRNAs in mutants relative to wt shows that DCL4 is required for 21 nt siRNA, DCL2 for 22 nt siRNA, and DCL3 for 24 nt siRNA accumulation (Figure 2A). Interestingly, the elimination of 21 nt viral siRNAs and endogenous ta-siRNAs (siR255) in dcl4 was associated with an increase in 22 and 24 nt species. Similarly, the elimination of 24 nt viral siRNAs and reduction in the 24 nt ra-siRNA (siR1003) in dcl3 correlated with an increase in 22 and 21 nt species. This indicates that other DCLs compensate for deficiencies in a specific DCL during the production of viral siRNAs. Similar observations were reported previously for endogenous sRNAs (11). In contrast, accumulation of viral siRNAs did not decrease in rdr2 or rdr6 (Figure 2A), indicating that viral siRNAs do not arise from known RDR-dependent endogenous RNA silencing pathways.
Levels of ChlI mRNA were reduced in all the mutant and wt plants infected with CaLCuV::Chl (Figure 1C and D). Therefore Chl-VIGS at the whole plant level depends neither on a single, specific size class of viral siRNA nor on RDR6 per se. Nevertheless, VIGS knock-down of ChlI mRNA was less potent in dcl4 and rdr6 (ca. 50%) than in wt, dcl2, dcl3 and rdr2 (Figure 1D); this correlated with the absence of chlorata whitening in newly formed tissues (Figure 1A). Similar results were obtained when non-inoculated inner rosette leaves of CaLCuV::Chl-infected dcl4 and rdr6 plants were analyzed for the ChlI mRNA levels (Supplementary Figure S2). Either the marginal enrichment in ChlI mRNA levels in these two mutants is sufficient to exceed the threshold level of CHLI required for chlorophyll synthesis in new growth or, more likely, the DCL4/RDR6-dependent silencing process underlying extensive spread of the chlorata phenotype initiates at very early stages of leaf development (see Discussion). We conclude that although DCL4 and RDR6 are not absolutely required for VIGS-mediated knockdown of ChlI mRNA in infected tissues, these genes play a special role in the establishment and/or maintenance of extensive VIGS in newly emerging leaves absent before inoculation.
Similar to our results for CaLCuV, CaMV-infected dcl and rdr plants exhibited no drastic effect on viral symptoms or titers (Supplementary Figure S1). Also, the biogenesis of 24 and 22 nt CaMV siRNAs required DCL3 and DCL2, respectively (Figure 3A). Production of CaMV 21 nt siRNAs was not abolished in dcl4 (Figure 3A), suggesting that unlike CaLCuV, a different DCL (e.g. DCL1) produces the remaining fraction of CaMV 21-nt siRNAs. The ratio between 21 and 24 nt siRNAs varied for different CaMV regions tested, with the lowest ratio for the TAV coding region. Furthermore, the effect of dcl4 on the relative accumulation of 21 nt viral siRNAs was more pronounced for TAV coding and L2 non-coding regions than for the L1 non-coding region (Figure 3). Presently, it is unclear why relative DCL specificities vary for different viral regions. An intriguing possibility is that structural features of viral dsRNA and stem–loop substrates derived from those regions (see Discussion) determine the DCL specificity.
The mutants rdr2 and rdr6 showed no deficiency for CaMV siRNA accumulation, indicating that like for CaLCuV, RDR2- and RDR6-dependent pathways are not required (Figure 3A). We obtained further evidence distinguishing viral siRNA biogenesis from endogenous pathways using loss-of-function mutants of POL IVa and AGO4 (components of the ra-siRNA pathway). Although accumulation of siR1003 was abolished in nrpd1a-3, a null allele of the POL IVa largest subunit (14), no effect was found on viral symptoms, titers, siRNA production or the spread of Chl-VIGS (Supplementary Figure S3). Thus POL IVa is not part of the plant response to DNA viruses associated with 24 nt viral siRNAs. In ago4 plants [La-er background (16)], a reduced accumulation of CaLCuV and CaMV siRNAs was observed, which correlated with a similar reduction in siR1003 levels (Supplementary Figure S3C and D). However, viral symptoms and titers were similar to those of the wild type control plants (Supplementary Figure S3A and B). Note that in CaLCuV::Chl-infected ago4 and La-er plants the Chl-VIGS phenotype of inner rosette was generally less developed (Supplementary Figure S3A), possibly because they bolted soon after inoculation.
DNA virus infection, viral siRNA biogenesis and VIGS in mutants for miRNA biogenesis
To test whether the miRNA pathway is important for production of siRNAs from DNA viruses, we inoculated the mutants dcl1-8, dcl1-9, hyl1-2 and hen1-5, which are all deficient for miRNA biogenesis (43,44).
Due to these mutants' retarded, abnormal development, it was difficult to evaluate virus susceptibility and symptoms. To obtain comparable sized plants, mutants were inoculated one month later than wt plants. Symptom development caused by both viruses was delayed in the mutants. CaLCuV::Chl-infected hen1-5 showed decreased Chl-VIGS whitening in emerging leaves compared to wt, dcl1-8, dcl1-9 and hyl1-2 (Supplementary Figure S4A). Two months post-inoculation, viral titers in all miRNA biogenesis mutants were comparable to wt levels for CaLCuV (data not shown) and CaMV (Figure S4B). The accumulation of 21, 22 and 24 nt CaLCuV siRNAs of both polarities was similar in dcl1-8, dcl1-9, hyl1-2 and wt plants (Figure 2B), whereas the controls siR255 and miR173 were undetectable in dcl1-8 and dcl1-9 and reduced in hyl1-2. Therefore, HYL1 and DCL1 are not required for biogenesis of the CaLCuV siRNAs tested.
During CaMV infection the relative accumulation of 21 nt viral siRNAs was strongly reduced in dcl1-9 and less strongly in dcl1-8 (Figure 3B). The fact that dcl1-8 carries a point mutation in the DCL1 helicase domain, while dcl1-9 has a T-DNA disruption in the DCL1 dsRNA binding domain (43) suggests that the latter domain is more important for 21 nt viral siRNA biogenesis. The truncated DCL1 protein likely expressed in dcl1-9 has been implicated in production of aberrant miRNAs (5). This defective DCL1 as well as DCL4 (see above) might therefore contribute together to production of the remaining fraction of viral 21 nt siRNAs in this mutant background. A reduced accumulation of 21 nt CaMV siRNA was also observed in hyl1-2 (Figure 3B), indicating that the DCL1 binding partner HYL1 (6) facilitates the biogenesis of a fraction of 21 nt CaMV siRNAs.
In hen1-5, the patterns of CaLCuV and CaMV siRNAs were slightly retarded, with additional ∼23–25 nt bands detected (Figures 2B and 3B). All these viral species, as well as endogenous sRNAs were sensitive to β-elimination in hen1-5, whereas those from wt plants were resistant (Supplementary Figure S5 and data not shown). Thus all size-clases of siRNAs derived from both plant DNA viruses are methylated by HEN1. Since CaLCuV::Chl-infected hen1-5 exhibited little chlorata whitening in newly formed tissues (Figure S4A), HEN1-mediated methylation of viral siRNAs is important for extensive VIGS in emerging leaves.
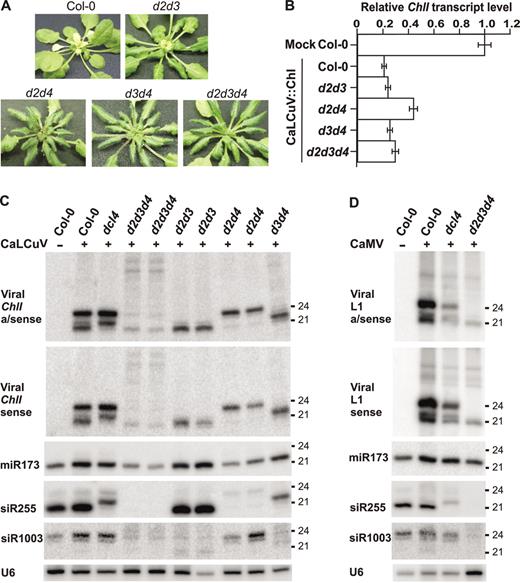
Effect of double and triple dcl-mutants on viral siRNA production and DNA-VIGS
Redundancy and compensation in the DCL family was investigated by examining the effect of double mutants d2d3, d2d4, d3d4 and the triple mutant d2d3d4 on virus infection (see Materials and Methods concerning crosses). CaLCuV::Chl inoculation resulted in normal viral symptom development; viral titers were similar in wt and mutant plants (data not shown). However, while d2d3 exhibited chlorata whitening of emerging leaves (Figure 4A), the mutant combinations including dcl4 did not. Nevertheless, all dcl-mutant combinations showed knock-down of endogenous ChlI mRNA levels (Figure 4B), which correlated with the presence of viral ChlI siRNAs (Figure 4C). As expected from our analysis of individual dcl-mutants, d2d3, d3d4 and d2d4 accumulated single size classes of 21, 22 or 24 nt viral siRNAs, respectively. Surprisingly, d2d3d4 still accumulated residual 21 nt viral siRNAs (Figure 4C). Apparently, in the absence of the other DCLs, DCL1 processes some CaLCuV dsRNAs. Notably, longer bands of both sense and antisense polarities, presumably viral dsRNA intermediates, were detected in d2d3d4 (Figure 4C). In the case of some RNA viruses, including Tobacco rattle virus (TRV) and Turnip crinkle virus (TCV) but not Cucumber mosaic virus (CMV), residual DCL1-derived viral sRNAs have been also observed (30,31).
Effect of double and triple dcl-mutations on DNA virus infection, siRNA biogenesis and VIGS. (A) Double mutants d2d3, d2d4, d3d4 and the triple mutant d2d3d4 1 month post-inoculation with CaLCuV::Chl. (B) Quantitative real-time RT–PCR for CaLCuV::Chl-infected double and triple mutants, performed as in Figure 1D. (C) LMW RNA blot analysis of Col-0 mock and CaLCuV::Chl-infected Col-0, dcl4, dcl-triple and double mutants. Two RNA sample pools are shown for infected d2d3d4, d2d3 and d2d4. (D) LMW RNA blot analysis of Col-0 mock and CaMV-infected Col-0, dcl4, d2d3d4 mutants. U6 signal is a loading control.
Taken together our results indicate that each DCL individually can mediate VIGS in infected tissues, which correlates with predominant accumulation of a corresponding single size-class of siRNA. Based on phenotypic observations for a different endogenous VIGS marker gene and an RNA virus TRV vector, Deleris et al. (30) concluded that, despite the presence of TRV-derived 24 nt siRNAs, RNA-VIGS does not occur in their Arabidopsis double mutant dcl2 dcl4. Our DNA-VIGS results show that when DCL3-dependent 24 nt viral siRNAs are the predominant species produced (i.e. in our d2d4), knock-down of ChlI mRNA still occurs (Figure 4B). This might illustrate a difference between RNA-VIGS and DNA-VIGS phenomena. However, we cannot formally exclude that trace amounts of DCL1-dependent siRNAs produced in d2d4 (Figure 4C) also contribute to DNA-VIGS, as they are capable of triggering ChlI mRNA knock-down in the triple mutant d2d3d4 (Figure 4B and 4C).
Like for CaLCuV, the triple mutant d2d3d4 did not exhibit increased susceptibility to CaMV compared to wt: symptom development and viral DNA titers were comparable (data not shown). Furthermore, d2d3d4 accumulated high levels of 21 nt CaMV siRNAs (Figure 4D), confirming the key role of DCL1 in their biogenesis.
CaMV interferes with dsRNA processing
In CaMV-infected wt plants, but not in CaLCuV-infected plants, ladders of longer RNA bands were detected. We observed this with viral and endogenous ta-siRNA probes (Figure 5). The band patterns were similar for sense and antisense polarities, revealing that they are likely dsRNA intermediates processed from longer precursors. ta-siRNA255 is generated from TAS 1a/b/c primary transcript(s) by miR173-mediated cleavage followed by RDR6 catalyzed second strand synthesis and DCL4 processing of the resulting dsRNA (10–13). CaMV infection might hinder DCL4-mediated processing of these precursors. Supporting this idea, no siR255-specific bands were detected in CaMV-infected rdr6, while their pattern changed in dcl4 but not in other dcl mutants (Figure 5B and data not shown). Consistent with impaired DCL4 processivity is the observed involvement of DCL1 in producing 21 nt CaMV siRNAs (Figure 3B).
Accumulation of endogenous and viral dsRNA in CaMV-infected plants. (A) Blot hybridization analysis of total RNA (30 μg/lane) from Col-0/La-er mock and CaMV-infected plants with probes for trans-acting siR255 and its duplex cognate siR255∗. (B) RNA (10 μg/lane) analyzed as in part A from CaMV-infected rdr6, rdr2 and dcl4 plants. (C) Membrane from part A was successively hybridized with sense and antisense probes for the CaMV transcript leader region (L1 and L2) and a coding region (TAV). Viral antisense 21, 22 and 24 nt siRNA strands of L1 and L2 regions migrate faster than sense strands due to their lower molecular weight (see Supplementary Table S1 for purine/pyrimidine contents).
The long antisense RNAs we detected for different regions of the CaMV genome (Figure 5C) reflect a considerable amount of antisense transcription of CaMV, which was not yet appreciated. In contrast to the aforementioned TAS precursors, accumulation of sense and antisense viral long RNA was RDR6-independent (data not shown).
Silencing and its suppression during RNA tobamovirus infection
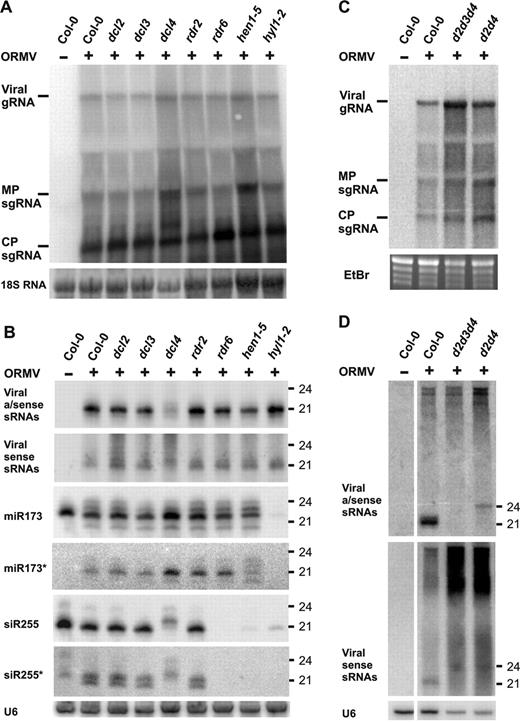
To compare our results for nuclear DNA viruses with a cytoplasmic RNA virus, we inoculated the Arabidopsis silencing mutants with ORMV. None of the mutants tested showed a dramatic difference in ORMV symptoms (data not shown). However, 22 days post-inoculation, viral RNA was significantly elevated in dcl4, d2d4 and d2d3d4 compared to wt or other mutants that did not affect DCL4 (Figure 6A and C). In contrast to the DNA viruses, ORMV siRNAs were predominantly of the 21 nt size class (Figure 6B and D), which was abolished in dcl4, d2d4 and d2d3d4, but not other mutants (Figure 6B and D). In dcl4, 22 nt siRNAs appeared, mostly of viral sense polarity (Figure 6B), while in d2d4 a 24 nt siRNA signal was detected (Figure 6D). In d2d3d4, nearly all siRNAs were abolished. Thus, DCL4 is the major dicer generating ORMV siRNAs. In its absence, DCL2 acts to generate 22 nt viral siRNAs, and when both DCL4 and DCL2 are absent, DCL3 generates 24 nt viral siRNAs. This compensation effect was reflected in viral long RNA titers, which were inversely correlated with siRNAs levels in the respective dcl mutants. RDR2, RDR6 and HYL1 do not play a role in ORMV siRNA biogenesis (Figure 6). Most recently, the primacy of DCL4 in producing viral siRNAs was demonstrated for infections with two different RNA viruses (30,31).
Molecular analysis of ORMV infection in wt and RNA silencing mutant backgrounds. (A) RNA blot hybridization was performed (as in Figure 2) to detect ORMV genomic RNA (gRNA), MP and CP subgenomic RNA (sgRNA) in infected wt, dcl, rdr, hen1-5 and hyl1-2 plants. 18S RNA probe hybridization is a loading control. (B) LMW RNA blot analysis for plants in part A, using viral sense, antisense and endogenous control probes (see Supplementary Table S1 for probe details). U6 signal is a loading control. (C) Blot analysis of viral RNA in infected Col-0, d2d3d4 and d2d4 plants. EtBr staining is a loading control. (D) LMW RNA blot analysis of RNA in part C, hybridized with viral sense and antisense probes. U6 signal is a loading control.
We showed previously that viral and endogenous sRNAs are sensitive to β-elimination in ORMV-infected wt plants (19). This suggests that ORMV suppresses HEN1 activity. Here we analyzed ORMV-infected silencing mutants, looking for changes in accumulation and modification of viral siRNAs, endogenous sRNAs (miRNA and ta-siRNA) and their passenger strands (miRNA∗ and siRNA∗). In contrast to CaMV-, CaLCuV- and non-infected plants, miR173∗ (21 nt) and siR255∗ (20 and 21 nt) species accumulated in ORMV-infected plants (Figures 6C and Supplementary Figure S5). Using the β-elimination test comparing wt and hen1-5 plants, we found that endogenous sRNA passenger (∗) strands, a fraction of corresponding sRNAs and most ORMV siRNAs were not methylated (Supplementary Figure S5). We conclude that unmethylated sRNA duplexes over-accumulate in ORMV-infected cells.
Upon ORMV infection aberrant ∼20, 23 and 24 nt miR173 species accumulate (Figure 6C) and are sensitive to β-elimination (Supplementary Figure S5). Moreover, miR173∗ patterns resembled those of miR173, although they migrated faster on the gel (Figure 6C). This observation fits the predicted 21 nt length of miR173∗ (one nt shorter than miR173). Thus, ORMV infection phenocopies HEN1 deficiency, where unmethylated miRNAs are subject to both partial degradation and oligourydilation (21), although the latter effect was not detectable for every type of sRNA (Figures 6C and Supplementary Figure S5 and data not shown).
DISCUSSION
Viral siRNA biogenesis varies between plant viruses and is primarily determined by the DCL step
Our analysis of viral siRNAs in Arabidopsis RNA silencing mutants shows that both gemini- and pararetro-viruses—two families of nuclear DNA viruses—are targeted by all four DCL activities. This process results in 21, 22 and 24 nt siRNA production from viral coding and non-coding regions, but the relative contributions of DCL4 and DCL1 differ between CaLCuV and CaMV. In contrast, the cytoplasmic tobamovirus ORMV is primarily targeted by DCL4, resulting in production of predominantly 21 nt viral siRNAs. Thus, the three viruses are differentially targeted by subsets of the four plant DCLs.
We observed no significant contribution of the plant RNA polymerases RDR6, RDR2 or POL IVa to viral siRNA biogenesis. Endogenous 21 nt ta-siRNA biogenesis requires RDR6 to produce dsRNA for DCL4 and 24 nt ra-siRNA biogenesis requires POL IVa and RDR2 to produce dsRNA for DCL3 (4,15). The equivalent size-classes of viral siRNAs accumulate in the same DCL-determined manner without utilizing the upstream part of the endogenous pathways. Taken together with the findings of Gasciolli et al. (11) and Xie et al. (12), we conclude that Arabidopsis DCLs are versatile size-specific enzymes, acting on many different dsRNA substrates present in plant cells.
Our results do not exclude involvement of POL IVb (47), or RDRs 1, 3a, 3b and 3c (8) in viral siRNA production. However, POL IVb appears to be involved in maintaining heterochromatin at certain genomic repeats rather than ra-siRNA biogenesis (15,47). RDR1, although inducible by RNA viruses, is not required for the biogenesis of siRNAs derived from RNA viruses (30,48), the two DNA viruses (our unpublished data) or any endogenous loci tested so far (29). To date, there is no evidence that RDR3a/b/c have a function.
The three size-classes of DNA viral siRNAs detected here are probably produced from dsRNA rather than from hairpin precursors, since sense and antisense strands accumulate for each size class. However, certain hairpin-like regions of viral transcripts might be recognized as quasi-miRNA precursors by DCL1. Interestingly, two of the three CaMV regions we probed (L1 and L2) are part of a 450 nt long, branched stem–loop structure within the 35S RNA leader (46). Whether plant DNA viruses, like some of their animal counterparts (49,50), code for true miRNAs is still an open question.
DCL1 can process viral dsRNA
Our results suggest that DCL1, thought to mainly process hairpins in the miRNA pathway, also processes long dsRNA. We observed 21 nt siRNAs from CaLCuV in the triple mutant lacking DCL2, DCL3 and DCL4 (Figure 4C) and DCL1-dependent processing of certain CaMV dsRNAs to 21 nt siRNAs (Figure 3). For CaLCuV, DCL1-mediated processing of viral dsRNA is inefficient, because presumed intermediates accumulate in addition to 21 nt siRNAs in infected d2d3d4 plants. We hypothesize that DCL1 evolved to excise miRNA/miRNA∗ duplexes from hairpins, and lacks the processivity required to efficiently digest long dsRNA. Thus, one would expect DCL1 to be out-competed by DCL4 for siRNA production.
As mentioned above, DCL1-dependent viral siRNAs are not sufficient to mediate establishment of total DNA-VIGS in newly emerging leaves. This distinction from DCL4 products may be functionally important, because miRNAs produced by DCL1 are thought to act in a cell-autonomous manner to regulate plant development. Since involvement of DCL1 in endogenous siRNA production was recently reported (51,52), it would be interesting to test whether all such DCL1-dependent sRNAs act locally to the cells that produce them.
dsRNA accumulation during DNA virus infection
While RNA viruses are obliged to produce sense and antisense copies of their genome to replicate, only sense transcripts are required for replication of DNA viruses. In contrast to RNA viruses, DNA viruses do not encode an RDR that could theoretically complement host deficiencies for silencing-related RDRs. We show that both gemini- and pararetrovirus infections give rise to sense and antisense transcripts, which might form dsRNA precursors for viral siRNAs. To account for their production without POL IV and RDR activities we propose that POL II-driven transcription on circular viral genomes could produce overlapping sense/antisense transcripts. Indeed, bidirectional POL II promoters in geminiviruses normally generate converging left- and rightward transcripts (53,54). Thus, viral transcripts could extend beyond their overlapping polyadenylation signals prior to cleavage and polyadenylation. Degradation products of read-through transcripts have been detected for the geminiviral promoter region (54). No bi-directional transcription has been reported for pararetroviruses, but we speculate that the strong CaMV enhancer of the 35S and 19S RNA promoters (55) can also drive POL II transcription in the antisense direction. Bidirectional transcription driven by the CaMV 35S core promoter was documented for a reporter system in Arabidopsis (56).
Numerous recent reports indicate that large parts of animal and plant genomes are transcribed in both orientations (57). Although the biological significance of antisense transcription is still unclear, it could serve as a genome surveillance mechanism to silence endogenous DNA repeats and multiple circular minichromosomes generated by DNA virus infection.
A specific role for DCL4 and RDR6 in virus induced gene silencing
Using the CaLCuV::Chl vector, we demonstrate that establishment and/or maintenance of extensive VIGS in newly growing tissues requires DCL4, HEN1 and RDR6. DCL1, DCL2 and DCL3 present in combinations or individually are sufficient to support VIGS-mediated knockdown of ChlI transcript in the absence of DCL4, but not sufficient to establish the indicative chlorata phenotype in newly emerging leaves.
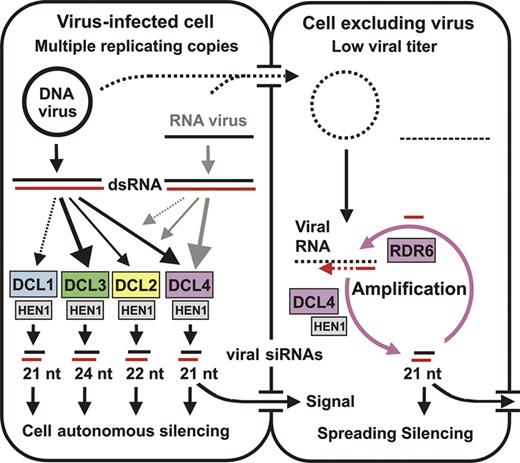
Our model to explain these observations (Figure 7) is that DCL4-dependent 21 nt ChlI siRNAs of viral origin spread into the shoot apical meristem, from which viruses are normally excluded, to trigger ChlI silencing in newly formed structures. Although Chl-VIGS in leaves formed before inoculation is associated with accumulation of three size classes of viral siRNAs, individual size classes are sufficient for ChlI mRNA knock-down. Indeed, strong accumulation of DCL2-dependent 22 nt siRNAs is correlated with reduced ChlI transcript levels in the d3d4 mutant. Similar results were obtained for DCL3-dependent 24 nt siRNA accumulation in d2d4, and for DCL1-dependent 21 nt siRNA accumulation in d2d3d4. Nonetheless, extensive VIGS did not occur in emerging leaves of these mutant plants deficient in DCL4 activity. Therefore, we suggest that DCL4-dependent 21 nt siRNAs are transported to the shoot apical meristem where they act as primers for RDR6 coupled to DCL4 in a signal amplification loop (Figure 7) and guide RISC-mediated degradation of target mRNAs. Both processes would require methylation of the 21 nt siRNAs mediated by HEN1. Alternatively, DCL4 and RDR6 might act only in the meristem to receive and amplify a silencing signal.
Model for VIGS and anti-viral defense based on RNA silencing. Two adjoining plant cells are shown schematically. The initially infected cell (left) contains high titers of DNA or RNA virus. Double-stranded RNA (dsRNA) arises from viral genomes independently of known silencing-related host RNA polymerases. Arrows connecting dsRNA to DCL enzymes represent the relative contribution of each DCL to viral siRNA biogenesis. For the DNA virus, every DCL digests the dsRNA into distinct size classes of siRNAs, with DCL3, DCL4 and DCL2 being favored (in that order). For RNA viruses, DCL4 is most important, with DCL2 and DCL3 compensating DCL4-deficiency (e.g. due to mutation or viral suppression). The 21 nt DCL4 product is potentially the signal required for VIGS spread. Both infectious nucleic acids and viral siRNAs move into the right-hand cell. However, the viral titer remains low, because DCL4 and RDR6 amplify incoming siRNA signal and digest viral transcripts. In this manner, VIGS would spread into meristematic tissues from which viruses are normally excluded. Viral siRNAs are stabilized by HEN1-mediated methylation.
Dunoyer et al. (24) concluded that cell-to-cell spread of silencing triggered by inverted-repeat transgenes requires DCL4 and is correlated with the accumulation of 21 nt but not 24 nt siRNAs. They proposed that primary 21 nt siRNAs are short-range silencing signals and that long-range silencing signals are secondary 21 nt siRNAs generated by RDR6-mediated amplification. Our findings support this hypothesis, extending it to the spread of VIGS. This would suggest a mechanism for the exclusion of RNA viruses from the shoot apical meristem. Earlier work established a role for RDR6 in this exclusion (26).
Compensation between DCL4-, DCL3- and DCL2-dependent viral siRNAs
The most abundant class of siRNAs derived from CaLCuV and CaMV is 24 nt long. Knocking out DCL3 eliminated 24 nt viral siRNAs without affecting viral DNA levels or VIGS. This is probably because DCL4 and DCL2 products (21 and 22 nt siRNAs) can mediate RNAi-like silencing in Arabidopsis (24,31). However, CaLCuV::Chl infection of the double mutant d2d4 still resulted in ChlI transcript knock-down, although mainly 24 nt viral ChlI siRNAs remained. Potentially, DCL3-dependent siRNAs are sufficient to target transcripts for degradation. This idea is reinforced by the higher accumulation of ORMV genomic RNA in d2d3d4 than in d2d4. Additionally, the 24 nt species might target the ChlI genomic DNA coding region, resulting in transcriptional silencing.
Our results implicate DCL2 in production of 22 nt viral siRNAs. These species accumulate in wt but not dcl2 plants infected with both DNA viruses studied here. Based on the increased accumulation of DCL2-dependent 22 nt siRNA in dcl3, dcl4 and d3d4, DCL2 appears to gain access to substrates normally processed by others DCLs. An antiviral role of DCL2 was suggested by Xie et al. (29) based on increased susceptibility of dcl2 plants to the RNA carmovirus TCV, which correlates with the absence of TCV-derived siRNAs at the early stage of infection. Nonetheless, infection with RNA viruses including CMV, TuMV (29) and ORMV (this study) did not result in differences of viral titers or siRNA accumulation in dcl2 or other mutants, except dcl4 (this study). We did notice, however, that DCL2-dependent 22 nt ORMV siRNAs accumulate in dcl4.
Based on their analyses of RNA viruses, Deleris et al. (30) and Bouche et al. (31) concluded that DCL4 plays a key role in anti-viral defense and RNA-VIGS but, when DCL4 is mutated or inhibited by viral suppression, DCL2 takes over this role. Our results for the RNA tobamovirus confirm such a hierarchical action of DCL4 and DCL2 and further show that DCL3 can also limit viral RNA accumulation.
Viral strategies of silencing suppression
RNA viruses might circumvent complex DCL redundancies by suppressing RNA silencing downstream, at the shared HEN1 step. We show that ORMV infection interferes with HEN1-mediated methylation of viral siRNAs and endogenous sRNAs, including miRNAs and ta-siRNAs, but not 24 nt ra-siRNAs [this work and (19)]. Furthermore, miRNA∗ and ta-siRNA∗ strands over-accumulate during ORMV infection. The tobamovirus factor responsible for interference with HEN1 remains to be identified. Mechanistically, one of the viral proteins might bind sRNA duplexes, as was demonstrated for several viral suppressors (35,36), or inactivate HEN1 directly through a protein-protein interaction. Unmethylated sRNA duplexes appear to undergo partial degradation at their 2 nt overhangs that results in accumulation of shorter-sized sRNAs of both viral and endogenous origin. Additionally, they are also subject to 3′-end oligouridylation observed at least for the miR173/miR173∗ duplex (Figure 6C). In both cases, assembly of RISC and the spread of silencing might be compromised. The proposed RISC assembly defect is supported by Baumberger and Baulcombe (7) who found that siRNAs derived from three RNA viruses (including another tobamovirus) are not associated with the RISC slicer component AGO1. It would be interesting to test whether other viruses from the AGO1 study interfere with HEN1.
When expressed as transgenes in Arabidopsis, silencing suppressors including potyviral HC-Pro, tombusviral p19 and closteroviral p21, known to bind miRNA and siRNA duplexes in vitro (35), increase accumulation of miRNA∗ strands (38) and prevent miRNA methylation by HEN1 (58). If these suppressors also interfere with HEN1 activity during viral infection, then targeting HEN1 would be a general silencing suppression strategy.
Additionally, our results suggest that the pararetrovirus CaMV has evolved a suppressor that stabilizes dsRNA products of RDR6. Indeed, several abundant long dsRNA precursors of ta-siRNAs accumulate in CaMV-infected plants. Potentially, the putative suppressor impairs the processing of these long dsRNAs by DCL4. Suppression of DCL4 in CaMV-infected tissues would be consistent with our result that the major fraction of 21 nt CaMV siRNAs depends on DCL1; during CaLCuV and ORMV infection, DCL4 performs this function instead. No silencing suppressor encoded by plant pararetroviruses has been reported so far. Our findings provide the basis for identifying this factor and for using CaMV as a tool to study ta-siRNA biogenesis.
The silencing suppression strategies we propose for CaMV and ORMV do not appear to be exploited by CaLCuV. DCL4- and HEN1-dependent production of methylated 21 nt viral siRNAs is not impaired in CaLCuV infected Arabidopsis. Geminiviruses encode at least two types of silencing suppressor proteins, AC2 and AC4 (59,60). The latter specifically binds single-stranded 21 nt sRNA in vitro and miRNA in vivo (60). Since VIGS initiated by CaLCuV::Chl spreads efficiently into emerging leaves via the action of DCL4, HEN1 and RDR6, geminivirus suppressors of silencing might act downstream of methylated 21 nt siRNA production.
CONCLUDING REMARKS
By testing a similar panel of Arabidopsis mutants as used in Deleris et al. (30) and Bouche et al. (31) for three RNA viruses, we found that two DNA viruses trigger essentially the same battery of siRNA responses as RNA viruses. A notable difference, however, is that DNA virus infection results in a strong accumulation of DCL3-dependent 24 nt siRNAs, irrespective of the viral region probed. In contrast, the equivalent size-class of RNA virus-derived species is detectable only in mutants lacking DCL4 and DCL2 (30, 31 and this work, for ORMV), with the exception of a recombinant RNA virus TRV, which produced 24 nt species in wild type plants (30). This distinction probably reflects the fundamentally different lifecycle of DNA viruses, which includes a nuclear phase.
Furthermore, our work demonstrates that all four DCL activities produce specific size-classes of DNA virus-derived siRNAs that contribute to the targeting of homologous ChlI mRNA to varying degrees. Surprisingly, mutations in silencing-related genes did not result in hypersusceptibility to ORMV or DNA virus infection. This is in contrast to the work of Bouche et al., who reported enhanced symptoms in CMV-infected plants of their triple mutant dcl2 dcl3 dcl4 line (31). We conclude that DCL pathway redundancies, as well as viral suppression of silencing in wt plants, hide broader deficiency phenotypes for infection with the viruses used in our study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank James Carrington for providing dcl4-2 seeds, David Baulcombe for nrpd1a-3, Olivier Voinnet for rdr2-1, rdr6-15 and ago4-1, S. Schauer for dcl1-8 seeds, and Fernando Ponz for the ORMV clone. Rashid Akbergenov was instrumental in helping R.R. and P.V.S. with small RNA analysis. Claudia Kutter and Sina Heinrichs helped characterize mutant plant materials. Estelle Arn and Konstantina Boutsika assisted with viral infections and plant care. We thank Barbara Hohn for her fruitful discussions on the work and D. Baulcombe, J. Burgyan, O. Voinnet and V. Vance for exchange of information. We are grateful to Thomas Boller and Christian Körner for the lab space and infrastructure at the Institute of Botany and the Friedrich Miescher Institute for the office space to TH. This work was supported by the Swiss National Science Foundation, the Indo-Swiss Collaboration in Biotechnology, the Swiss Office for Education and Science as part of the EU Gene Silencing in Transgenic Plants Projects, and by the Novartis Research Foundation. Funding to pay the Open Access publication charges for this article was provided by the Friedrich Miescher Institute.
Conflict of interest statement. None declared.
REFERENCES




Comments