-
PDF
- Split View
-
Views
-
Cite
Cite
Yukihisa Katsumoto, Masako Fukuchi-Mizutani, Yuko Fukui, Filippa Brugliera, Timothy A. Holton, Mirko Karan, Noriko Nakamura, Keiko Yonekura-Sakakibara, Junichi Togami, Alix Pigeaire, Guo-Qing Tao, Narender S. Nehra, Chin-Yi Lu, Barry K. Dyson, Shinzo Tsuda, Toshihiko Ashikari, Takaaki Kusumi, John G. Mason, Yoshikazu Tanaka, Engineering of the Rose Flavonoid Biosynthetic Pathway Successfully Generated Blue-Hued Flowers Accumulating Delphinidin, Plant and Cell Physiology, Volume 48, Issue 11, November 2007, Pages 1589–1600, https://doi.org/10.1093/pcp/pcm131
Close - Share Icon Share
Abstract
Flower color is mainly determined by anthocyanins. Rosa hybrida lacks violet to blue flower varieties due to the absence of delphinidin-based anthocyanins, usually the major constituents of violet and blue flowers, because roses do not possess flavonoid 3′,5′-hydoxylase (F3′5′H), a key enzyme for delphinidin biosynthesis. Other factors such as the presence of co-pigments and the vacuolar pH also affect flower color. We analyzed the flavonoid composition of hundreds of rose cultivars and measured the pH of their petal juice in order to select hosts of genetic transformation that would be suitable for the exclusive accumulation of delphinidin and the resulting color change toward blue. Expression of the viola F3′5′H gene in some of the selected cultivars resulted in the accumulation of a high percentage of delphinidin (up to 95%) and a novel bluish flower color. For more exclusive and dominant accumulation of delphinidin irrespective of the hosts, we down-regulated the endogenous dihydroflavonol 4-reductase (DFR) gene and overexpressed the Iris×hollandica DFR gene in addition to the viola F3′5′H gene in a rose cultivar. The resultant roses exclusively accumulated delphinidin in the petals, and the flowers had blue hues not achieved by hybridization breeding. Moreover, the ability for exclusive accumulation of delphinidin was inherited by the next generations.
Introduction
Flower color plays important roles in plant sexual hybridization by attracting pollinators, such as insects and birds. Flower color is also attractive to people and is an important characteristic of flowers from a floricultural viewpoint. Hybridization breeding has partially achieved the diversification of flower color of floricultural crops.
Plant species have adopted versatile and ingenious ways to exhibit flower color (Grotewold 2006, Tanaka and Brugliera 2006). Such ingenuity has been studied and understood in terms of chemistry, biochemistry and molecular biology (Forkmann and Heller 1999, Goto 1987, Tanaka and Brugliera 2006). Among various pigments in petals, anthocyanins, a class of flavonoids, are major constituents of flower color from orange/red to violet/blue. Their chemical structures primarily determine their color, i.e. the number of hydroxy groups on the B-ring and/or aromatic acyl moieties modifying anthocyanins increase, causing a bathochromic shift to blue (Honda and Saito 2002). The majority of violet/blue flowers contain delphinidin-based anthocyanins modified with one or more aromatic acyl moieties (Honda and Saito 2002). Anthocyanins change their color depending on the pH of the vacuole in which anthocyanins localize; their color is bluer in weakly acidic or neutral pH, and redder in acidic pH. Co-pigments, usually flavones and flavonols, cause a bathochromic shift of anthocyanins when they stack with anthocyanins (Goto and Kondo 1991). The formation of a complex with metal ions can give a blue color (Kondo et al. 1992, Yoshida et al. 2003, Shiono et al. 2005, Shoji et al. 2007).
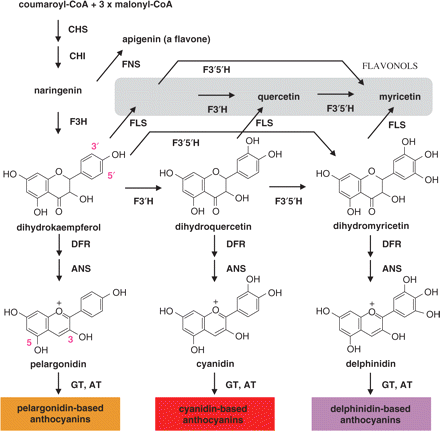
Among the factors influencing flower color, flavonoid/anthocyanin biosynthesis has been the most extensively studied. The pathway leading to anthocyanidin 3-glucoside is generally conserved among higher plant species (Fig. 1) (Forkmann and Heller 1999, Grotewold 2006, Tanaka and Brugliera 2006). Each plant species usually accumulates limited kinds of anthocyanins and exhibits limited flower color by the expression of a specific set of biosynthetic genes, the substrate specificity of key enzymes and/or the temporal and spatial regulation of the biosynthetic genes. Therefore, it is rare for a single species to have the entire spectrum of flower color, although floricultural breeders have always sought novel flower colors (Tanaka et al. 2005, Tanaka and Brugliera 2006). For example, roses, carnations and chrysanthemum, which are the most important floricultural crops, do not accumulate delphinidin-based anthocyanins. Thus, they lack violet/blue varieties. This is attributed to their deficiency of flavonoid 3′,5′-hydroxylase (F3′5′H), a key enzyme in the synthesis of delphinidin (Fig. 1) (Holton and Tanaka 1994)).
Generalized flavonoid biosynthetic pathway relevant to flower color. Native rose petals only accumulate pelargonodin and cyanidin-based anthocyanins, mainly pelargonidin and cyanidin 3,5-diglucoside. Lack of delphinidin-based anthocynanins, which is attributed to deficiency of F3′5′H, has hampered the generation of rose flowers having blue and violet hues. The expression of a hetelorogous F3′5′H gene in rose is expected to generate delphinidin and, thus, a novel flower color with a blue hue. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; FLS, flavonol synthase; FNS, flavone synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; GT, anthocyanidin glucosyltransferase; AT, anthocyanin acyltransferase.
On the other hand, petunia and cymbidium lack brick red/orange varieties due to the lack of pelargonidin-based anthocyanins because their dihydroflavonol 4-reductases (DFRs) do not utilize dihydrokaempferol as a substrate (Forkmann and Heller 1999, Johnson et al. 1999). Some species that usually accumulate delphinidin-based anthocyanins and do not accumulate pelargonidin-based anthocyanins in their petals, such as iris and gentian, are likely to have DFRs with a similar substrate specificity to that of petunia DFR. Rose DFR can utilize dihydromyricetin as a substrate on the basis of feeding of dihydromyricetin to rose petals (Holton and Tanaka 1994).
Advances in molecular biology and plant transformation technology have made possible the engineering of an anthocyanin biosynthetic pathway and, thus, flower color by the overexpression of heterologous flavonoid biosynthetic genes and/or the down-regulation of endogenous genes in transgenic plants, including petunia, torenia and carnation, as reviewed by Tanaka (2006). In carnation, the overexpression of the F3′5′H gene alone was insufficient to convert the metabolic flux fully toward delphinidin biosynthesis. White carnation cultivars that specifically lacked the DFR gene were transformed with the petunia or viola F3′5′H gene in combination with the petunia DFR gene. As a result, violet carnations accumulating delphinidin-based anthocyanins were almost exclusively generated (Holton 1996, Fukui et al. 2003). These results indicate that it is possible to generate a novel flower color by changing the structure of anthocyanin, more specifically the number of hydroxyl groups on the B-ring, with genetic modification of the pathway. They also indicate that the selection of cultivars that have proper genetic background and flavonoid compositions and/or the artificial down-regulation of a competing endogenous pathway is necessary to obtain a desirable phenotype (Tanaka 2006, Tanaka and Brugliera 2006).
Roses are the most important flowers in today's global flower market and have been the center of attraction for consumers and breeders for hundreds of years. Modern roses (Rosa hybrida) have resulted from extensive hybridization of wild rose species and have various flower colors, except those in the violet to blue range (Vries et al. 1974, Holton and Tanaka 1994). Rose breeders have long endeavored to create blue roses, but their efforts have so far only led to pink and pale mauve-colored flowers. This is not surprising because rose petals contain pelargonidin/cyanidin 3-glucoside or 3,5-diglucoside (Biolley and Jay 1993, Mikanagi et al. 2000) and carotenoids (Vries et al. 1974). Some roses produce only small amounts of acylated anthocyanins, and roses do not produce flavones (Mikanagi et al. 2000), which are stronger co-pigments than flavonol (Yabuya et al. 1997). The vacuolar pH of rose petal epidermal cells is low (from pH 3.69 to 5.78) (Biolley and Jay 1993). Therefore, roses fundamentally lack the components required to yield violet/blue flowers.
More recently, some rose cultivars have been shown to contain a small amount of blue pigments (rosacyanin) that are derivatives of cyanidin (Fukui et al. 2006). Blue coloration in the cultivar ‘Rhapsody in Blue’ is caused by the accumulation of cyanin (cyanidin 3,5-O-di-glucoside) in anthocyanic vacuolar inclusions (AVIs) (Gonnet 2003). However, the molecular mechanisms of rosacyanin and AVIs is not yet understood, and their utilization to engineer blue/violet roses is not currently possible.
Although there may be multiple ways to create blue roses, we have chosen delphinidin production in rose petals because delphinidin biosynthesis is regulated by a single enzyme, F3′5′H, and its overexpression is unlikely to have a detrimental effect on plants. Delphinidin production presents a major breakthrough in the achievement of blue roses (Holton and Tanaka 1994). The expression of viola F3′5′H genes produced delphinidin (Brugliera et al. 2004). However, the presence of pelargonidin and cyanidin-based anthocyanins derived from the endogenous pathway prevented sufficient coloration toward blue. Unlike carnations, we were not able to obtain white rose cultivars that specifically lacked DFR activity.
Here, we analyzed the flavonoid composition of many rose cultivars in order to select cultivars which will exhibit a blue hue when delphinidin is accumulated. The overexpression of a viola F3′5′H gene resulted in the efficient accumulation of delphinidin and color changes with a novel blue hue in a few of the selected cultivars. Furthermore, the down-regulation of the rose DFR gene and overexpression of the iris DFR gene, as well as the overexpression of the viola F3′5′H gene, resulted in more efficient and exclusive delphinidin production and a bluer flower color. It is noteworthy that exclusive delphinidin production is heritable by the progeny, which should change rose breeding as far as flower color is concerned.
Results
Cultivar screening for engineering a flavonoid pathway
Hundreds of rose cultivars that claimed to be ‘blue’ and/or have names associated with ‘blue’ have been commercialized. Their flower color ranges are relatively blue among roses and in fact range from pink/red purple to gray/mauve. Some of these cultivars were subjected to flavonoid analysis and pH measurements, and the results are shown in Table 1. Their petals contained mainly cyanidin as an anthocyanidin, but not delphinidin. They contained a large amount of flavonols. Their bluish color may be due to a high flavonol/anthocyanidin ratio, and flavonols may function as co-pigments. Their higher pH than that of red roses may also contribute to their flower color (Table 1). These results are consistent with a previous report (Biolley and Jay 1993). Table 1 includes some red cultivars for comparison, which contain cyanidin and/or pelargonidin. They tend to have fewer flavonols and a lower pH. We recently discovered that some of these roses contain small amounts of novel cyanidin derivatives (rosacyanins) exhibiting a blue color (Fukui et al. 2006), but the degree of their contribution to flower color is not clear.
Flavonoid composition of commercial bluish rose varieties
| Cultivar . | Anthocyanidin (mg g−1)a . | Flavonol (mg g−1)a . | pH . | |||
|---|---|---|---|---|---|---|
| . | cya . | pel . | peo . | Q . | K . | . |
| ‘Blue’ varieties | ||||||
| Blue Bajou | 0.048 | 3.956 | 0.379 | 5.11 | ||
| Blue Heaven | 0.008 | 1.504 | 0.569 | 4.90 | ||
| Blue Moon | 0.049 | 1.341 | 0.119 | 5.25 | ||
| Delilah | 0.066 | 0.611 | 0.277 | 5.35 | ||
| Grey Pearl | 0.036 | 3.708 | 0.417 | 5.12 | ||
| Lavande | 0.109 | 1.020 | 0.195 | 4.70 | ||
| Madam Violet | 0.060 | 1.780 | 0.150 | 4.70 | ||
| Ondina | 0.019 | 2.056 | 1.043 | 4.99 | ||
| Rhapsody in Blue | 1.851 | 0.011 | 0.005 | 3.456 | 1.622 | 5.67 |
| Seiryu | 0.015 | 3.030 | 1.300 | 5.13 | ||
| Shocking Blue | 0.196 | 1.198 | 0.194 | 5.40 | ||
| Sterling Silver | 0.035 | 1.495 | 0.363 | 5.71 | ||
| Vol de Nuit | 0.317 | 0.003 | 2.661 | 0.316 | 4.90 | |
| Watarase | 0.056 | 3.048 | 0.124 | 5.07 | ||
| Pink-red varieties, etc. | ||||||
| Black Baccara | 4.043 | 0.007 | 0.705 | 0.119 | 4.58 | |
| First Red | 1.959 | 0.291 | 0.086 | 0.340 | 4.70 | |
| Jacaranda | 0.315 | 0.000 | 1.238 | 0.468 | 4.80 | |
| Kardinal | 1.898 | 0.125 | 0.156 | 0.296 | 4.40 | |
| Kiss | 0.006 | 0.038 | 0.028 | 1.381 | 4.02 | |
| Lambada | 0.036 | 0.164 | 0.014 | 0.466 | 4.40 | |
| Medeo | 0.004 | 0.004 | 0.028 | 2.323 | 4.40 | |
| Rote Rose | 3.480 | 0.034 | 0.238 | 0.285 | 4.37 | |
| Sonia | 0.060 | 0.300 | 0.006 | 0.650 | 4.85 | |
| Cultivar . | Anthocyanidin (mg g−1)a . | Flavonol (mg g−1)a . | pH . | |||
|---|---|---|---|---|---|---|
| . | cya . | pel . | peo . | Q . | K . | . |
| ‘Blue’ varieties | ||||||
| Blue Bajou | 0.048 | 3.956 | 0.379 | 5.11 | ||
| Blue Heaven | 0.008 | 1.504 | 0.569 | 4.90 | ||
| Blue Moon | 0.049 | 1.341 | 0.119 | 5.25 | ||
| Delilah | 0.066 | 0.611 | 0.277 | 5.35 | ||
| Grey Pearl | 0.036 | 3.708 | 0.417 | 5.12 | ||
| Lavande | 0.109 | 1.020 | 0.195 | 4.70 | ||
| Madam Violet | 0.060 | 1.780 | 0.150 | 4.70 | ||
| Ondina | 0.019 | 2.056 | 1.043 | 4.99 | ||
| Rhapsody in Blue | 1.851 | 0.011 | 0.005 | 3.456 | 1.622 | 5.67 |
| Seiryu | 0.015 | 3.030 | 1.300 | 5.13 | ||
| Shocking Blue | 0.196 | 1.198 | 0.194 | 5.40 | ||
| Sterling Silver | 0.035 | 1.495 | 0.363 | 5.71 | ||
| Vol de Nuit | 0.317 | 0.003 | 2.661 | 0.316 | 4.90 | |
| Watarase | 0.056 | 3.048 | 0.124 | 5.07 | ||
| Pink-red varieties, etc. | ||||||
| Black Baccara | 4.043 | 0.007 | 0.705 | 0.119 | 4.58 | |
| First Red | 1.959 | 0.291 | 0.086 | 0.340 | 4.70 | |
| Jacaranda | 0.315 | 0.000 | 1.238 | 0.468 | 4.80 | |
| Kardinal | 1.898 | 0.125 | 0.156 | 0.296 | 4.40 | |
| Kiss | 0.006 | 0.038 | 0.028 | 1.381 | 4.02 | |
| Lambada | 0.036 | 0.164 | 0.014 | 0.466 | 4.40 | |
| Medeo | 0.004 | 0.004 | 0.028 | 2.323 | 4.40 | |
| Rote Rose | 3.480 | 0.034 | 0.238 | 0.285 | 4.37 | |
| Sonia | 0.060 | 0.300 | 0.006 | 0.650 | 4.85 | |
cya, cyanidin; pel, pelargonidin; peo, peonidin; Q, quercetin; K, kaempferol.
a mg g−1 fresh petals.
Flavonoid composition of commercial bluish rose varieties
| Cultivar . | Anthocyanidin (mg g−1)a . | Flavonol (mg g−1)a . | pH . | |||
|---|---|---|---|---|---|---|
| . | cya . | pel . | peo . | Q . | K . | . |
| ‘Blue’ varieties | ||||||
| Blue Bajou | 0.048 | 3.956 | 0.379 | 5.11 | ||
| Blue Heaven | 0.008 | 1.504 | 0.569 | 4.90 | ||
| Blue Moon | 0.049 | 1.341 | 0.119 | 5.25 | ||
| Delilah | 0.066 | 0.611 | 0.277 | 5.35 | ||
| Grey Pearl | 0.036 | 3.708 | 0.417 | 5.12 | ||
| Lavande | 0.109 | 1.020 | 0.195 | 4.70 | ||
| Madam Violet | 0.060 | 1.780 | 0.150 | 4.70 | ||
| Ondina | 0.019 | 2.056 | 1.043 | 4.99 | ||
| Rhapsody in Blue | 1.851 | 0.011 | 0.005 | 3.456 | 1.622 | 5.67 |
| Seiryu | 0.015 | 3.030 | 1.300 | 5.13 | ||
| Shocking Blue | 0.196 | 1.198 | 0.194 | 5.40 | ||
| Sterling Silver | 0.035 | 1.495 | 0.363 | 5.71 | ||
| Vol de Nuit | 0.317 | 0.003 | 2.661 | 0.316 | 4.90 | |
| Watarase | 0.056 | 3.048 | 0.124 | 5.07 | ||
| Pink-red varieties, etc. | ||||||
| Black Baccara | 4.043 | 0.007 | 0.705 | 0.119 | 4.58 | |
| First Red | 1.959 | 0.291 | 0.086 | 0.340 | 4.70 | |
| Jacaranda | 0.315 | 0.000 | 1.238 | 0.468 | 4.80 | |
| Kardinal | 1.898 | 0.125 | 0.156 | 0.296 | 4.40 | |
| Kiss | 0.006 | 0.038 | 0.028 | 1.381 | 4.02 | |
| Lambada | 0.036 | 0.164 | 0.014 | 0.466 | 4.40 | |
| Medeo | 0.004 | 0.004 | 0.028 | 2.323 | 4.40 | |
| Rote Rose | 3.480 | 0.034 | 0.238 | 0.285 | 4.37 | |
| Sonia | 0.060 | 0.300 | 0.006 | 0.650 | 4.85 | |
| Cultivar . | Anthocyanidin (mg g−1)a . | Flavonol (mg g−1)a . | pH . | |||
|---|---|---|---|---|---|---|
| . | cya . | pel . | peo . | Q . | K . | . |
| ‘Blue’ varieties | ||||||
| Blue Bajou | 0.048 | 3.956 | 0.379 | 5.11 | ||
| Blue Heaven | 0.008 | 1.504 | 0.569 | 4.90 | ||
| Blue Moon | 0.049 | 1.341 | 0.119 | 5.25 | ||
| Delilah | 0.066 | 0.611 | 0.277 | 5.35 | ||
| Grey Pearl | 0.036 | 3.708 | 0.417 | 5.12 | ||
| Lavande | 0.109 | 1.020 | 0.195 | 4.70 | ||
| Madam Violet | 0.060 | 1.780 | 0.150 | 4.70 | ||
| Ondina | 0.019 | 2.056 | 1.043 | 4.99 | ||
| Rhapsody in Blue | 1.851 | 0.011 | 0.005 | 3.456 | 1.622 | 5.67 |
| Seiryu | 0.015 | 3.030 | 1.300 | 5.13 | ||
| Shocking Blue | 0.196 | 1.198 | 0.194 | 5.40 | ||
| Sterling Silver | 0.035 | 1.495 | 0.363 | 5.71 | ||
| Vol de Nuit | 0.317 | 0.003 | 2.661 | 0.316 | 4.90 | |
| Watarase | 0.056 | 3.048 | 0.124 | 5.07 | ||
| Pink-red varieties, etc. | ||||||
| Black Baccara | 4.043 | 0.007 | 0.705 | 0.119 | 4.58 | |
| First Red | 1.959 | 0.291 | 0.086 | 0.340 | 4.70 | |
| Jacaranda | 0.315 | 0.000 | 1.238 | 0.468 | 4.80 | |
| Kardinal | 1.898 | 0.125 | 0.156 | 0.296 | 4.40 | |
| Kiss | 0.006 | 0.038 | 0.028 | 1.381 | 4.02 | |
| Lambada | 0.036 | 0.164 | 0.014 | 0.466 | 4.40 | |
| Medeo | 0.004 | 0.004 | 0.028 | 2.323 | 4.40 | |
| Rote Rose | 3.480 | 0.034 | 0.238 | 0.285 | 4.37 | |
| Sonia | 0.060 | 0.300 | 0.006 | 0.650 | 4.85 | |
cya, cyanidin; pel, pelargonidin; peo, peonidin; Q, quercetin; K, kaempferol.
a mg g−1 fresh petals.
The criteria for the selection of possible host cultivars to achieve flower color modification toward blue by delphinidin production were: (i) they accumulated flavonols that were expected to be co-pigments; (ii) they had a higher vacuolar pH; (iii) ideally, they did not have F3′H activity; and (iv) they accumulated pelargonidin rather than cyanidin.
In order to choose cultivars to meet these criteria, several hundreds of rose flower cultivars were screened initially by visual observation; pink to mauve roses were mainly selected. Dark color cultivars and red cultivars were not selected, since they did not contain much flavonol and their pH tends to be low. Yellow and white cultivars were not selected either, as they are not able to accumulate anthocyanins in the petals. The bluish rose cultivars shown in Table 1 were not subjected to further study because of their low transformation frequencies or their low anthocyanin contents to achieve clear color change. The petals of 169 selected cultivars were harvested and subjected to flavonoid analysis and pH measurements. Some of the results are shown in Table 2 (see controls).
Flavonoid composition of Keisei rose varieties transformed with pSPB130
| Host . | Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | acyl (%) . | Flavonol (mg g−1)a . | pH . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | del . | cya . | pel . | . | . | M . | Q . | K . | . |
| WKS77 | Control | 0.000 | 1.103 | 0.005 | 0.0% | 0.0% | 0.000 | 1.513 | 0.303 | 5.05 | |
| pSPB130 | Average (22) | 0.600 | 1.343 | 0.007 | 32.4% | 10.2% | |||||
| Best line | 1.068 | 0.814 | 0.004 | 56.6% | 26.1% | 0.604 | 0.503 | 0.126 | |||
| WKS82 | Control | 0.000 | 0.124 | 0.000 | 0.0% | 0.0% | 0.000 | 1.598 | 0.081 | 5.46 | |
| pSPB130 | Average (52) | 0.055 | 0.038 | 0.000 | 54.4% | 9.0% | |||||
| Best line | 0.139 | 0.008 | 0.000 | 94.5% | 22.7% | 1.929 | 0.436 | 0.036 | |||
| WKS100 | Control | 0.000 | 0.066 | 0.000 | 0.0% | 0.0% | 0.000 | 1.248 | 0.368 | 5.34 | |
| pSPB130 | Average (71) | 0.044 | 0.044 | 0.000 | 45.6% | 11.2% | |||||
| Best line | 0.152 | 0.015 | 0.000 | 91.2% | 39.5% | 1.320 | 0.448 | 0.120 | |||
| WKS116 | Control | 0.000 | 0.017 | 0.000 | 0.0% | 0.0% | 0.000 | 1.725 | 0.636 | 5.29 | |
| pSPB130 | Average (40) | 0.040 | 0.012 | 0.000 | 73.2% | 8.7% | |||||
| Best line | 0.051 | 0.003 | 0.000 | 94.9% | 39.3% | 1.226 | 0.245 | 0.096 | |||
| WKS124 | Control | 0.000 | 0.022 | 0.045 | 0.0% | 0.0% | 0.000 | 0.192 | 2.011 | 4.85 | |
| pSPB130 | Average (16) | 0.728 | 0.098 | 0.021 | 82.3 % | 24.6% | |||||
| Best line | 1.017 | 0.058 | 0.000 | 94.6% | 44.2% | 1.161 | 0.140 | 0.262 | |||
| WKS140 | Control | 0.000 | 0.075 | 0.000 | 0.0% | 0.0% | 0.000 | 2.412 | 0.271 | 5.25 | |
| pSPB130 | Average (49) | 0.077 | 0.063 | 0.000 | 47.3% | 11.4% | |||||
| Best line | 0.114 | 0.007 | 0.000 | 94.1% | 33.6% | 0.689 | 0.136 | 0.031 | |||
| Host . | Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | acyl (%) . | Flavonol (mg g−1)a . | pH . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | del . | cya . | pel . | . | . | M . | Q . | K . | . |
| WKS77 | Control | 0.000 | 1.103 | 0.005 | 0.0% | 0.0% | 0.000 | 1.513 | 0.303 | 5.05 | |
| pSPB130 | Average (22) | 0.600 | 1.343 | 0.007 | 32.4% | 10.2% | |||||
| Best line | 1.068 | 0.814 | 0.004 | 56.6% | 26.1% | 0.604 | 0.503 | 0.126 | |||
| WKS82 | Control | 0.000 | 0.124 | 0.000 | 0.0% | 0.0% | 0.000 | 1.598 | 0.081 | 5.46 | |
| pSPB130 | Average (52) | 0.055 | 0.038 | 0.000 | 54.4% | 9.0% | |||||
| Best line | 0.139 | 0.008 | 0.000 | 94.5% | 22.7% | 1.929 | 0.436 | 0.036 | |||
| WKS100 | Control | 0.000 | 0.066 | 0.000 | 0.0% | 0.0% | 0.000 | 1.248 | 0.368 | 5.34 | |
| pSPB130 | Average (71) | 0.044 | 0.044 | 0.000 | 45.6% | 11.2% | |||||
| Best line | 0.152 | 0.015 | 0.000 | 91.2% | 39.5% | 1.320 | 0.448 | 0.120 | |||
| WKS116 | Control | 0.000 | 0.017 | 0.000 | 0.0% | 0.0% | 0.000 | 1.725 | 0.636 | 5.29 | |
| pSPB130 | Average (40) | 0.040 | 0.012 | 0.000 | 73.2% | 8.7% | |||||
| Best line | 0.051 | 0.003 | 0.000 | 94.9% | 39.3% | 1.226 | 0.245 | 0.096 | |||
| WKS124 | Control | 0.000 | 0.022 | 0.045 | 0.0% | 0.0% | 0.000 | 0.192 | 2.011 | 4.85 | |
| pSPB130 | Average (16) | 0.728 | 0.098 | 0.021 | 82.3 % | 24.6% | |||||
| Best line | 1.017 | 0.058 | 0.000 | 94.6% | 44.2% | 1.161 | 0.140 | 0.262 | |||
| WKS140 | Control | 0.000 | 0.075 | 0.000 | 0.0% | 0.0% | 0.000 | 2.412 | 0.271 | 5.25 | |
| pSPB130 | Average (49) | 0.077 | 0.063 | 0.000 | 47.3% | 11.4% | |||||
| Best line | 0.114 | 0.007 | 0.000 | 94.1% | 33.6% | 0.689 | 0.136 | 0.031 | |||
del, delphinidin; cya, cyanidin; pel, pelargonidin; del (%), delphinidin/total anthocyanidin; acyl (%), acylated anthocyanin/total anthocyanin; M, myricetin; Q, quercetin; K, kaempferol; best line, a line producing the highest percentage of delphinidin and exhibiting clear color change.
a mg g−1 fresh petals.
Flavonoid composition of Keisei rose varieties transformed with pSPB130
| Host . | Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | acyl (%) . | Flavonol (mg g−1)a . | pH . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | del . | cya . | pel . | . | . | M . | Q . | K . | . |
| WKS77 | Control | 0.000 | 1.103 | 0.005 | 0.0% | 0.0% | 0.000 | 1.513 | 0.303 | 5.05 | |
| pSPB130 | Average (22) | 0.600 | 1.343 | 0.007 | 32.4% | 10.2% | |||||
| Best line | 1.068 | 0.814 | 0.004 | 56.6% | 26.1% | 0.604 | 0.503 | 0.126 | |||
| WKS82 | Control | 0.000 | 0.124 | 0.000 | 0.0% | 0.0% | 0.000 | 1.598 | 0.081 | 5.46 | |
| pSPB130 | Average (52) | 0.055 | 0.038 | 0.000 | 54.4% | 9.0% | |||||
| Best line | 0.139 | 0.008 | 0.000 | 94.5% | 22.7% | 1.929 | 0.436 | 0.036 | |||
| WKS100 | Control | 0.000 | 0.066 | 0.000 | 0.0% | 0.0% | 0.000 | 1.248 | 0.368 | 5.34 | |
| pSPB130 | Average (71) | 0.044 | 0.044 | 0.000 | 45.6% | 11.2% | |||||
| Best line | 0.152 | 0.015 | 0.000 | 91.2% | 39.5% | 1.320 | 0.448 | 0.120 | |||
| WKS116 | Control | 0.000 | 0.017 | 0.000 | 0.0% | 0.0% | 0.000 | 1.725 | 0.636 | 5.29 | |
| pSPB130 | Average (40) | 0.040 | 0.012 | 0.000 | 73.2% | 8.7% | |||||
| Best line | 0.051 | 0.003 | 0.000 | 94.9% | 39.3% | 1.226 | 0.245 | 0.096 | |||
| WKS124 | Control | 0.000 | 0.022 | 0.045 | 0.0% | 0.0% | 0.000 | 0.192 | 2.011 | 4.85 | |
| pSPB130 | Average (16) | 0.728 | 0.098 | 0.021 | 82.3 % | 24.6% | |||||
| Best line | 1.017 | 0.058 | 0.000 | 94.6% | 44.2% | 1.161 | 0.140 | 0.262 | |||
| WKS140 | Control | 0.000 | 0.075 | 0.000 | 0.0% | 0.0% | 0.000 | 2.412 | 0.271 | 5.25 | |
| pSPB130 | Average (49) | 0.077 | 0.063 | 0.000 | 47.3% | 11.4% | |||||
| Best line | 0.114 | 0.007 | 0.000 | 94.1% | 33.6% | 0.689 | 0.136 | 0.031 | |||
| Host . | Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | acyl (%) . | Flavonol (mg g−1)a . | pH . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | del . | cya . | pel . | . | . | M . | Q . | K . | . |
| WKS77 | Control | 0.000 | 1.103 | 0.005 | 0.0% | 0.0% | 0.000 | 1.513 | 0.303 | 5.05 | |
| pSPB130 | Average (22) | 0.600 | 1.343 | 0.007 | 32.4% | 10.2% | |||||
| Best line | 1.068 | 0.814 | 0.004 | 56.6% | 26.1% | 0.604 | 0.503 | 0.126 | |||
| WKS82 | Control | 0.000 | 0.124 | 0.000 | 0.0% | 0.0% | 0.000 | 1.598 | 0.081 | 5.46 | |
| pSPB130 | Average (52) | 0.055 | 0.038 | 0.000 | 54.4% | 9.0% | |||||
| Best line | 0.139 | 0.008 | 0.000 | 94.5% | 22.7% | 1.929 | 0.436 | 0.036 | |||
| WKS100 | Control | 0.000 | 0.066 | 0.000 | 0.0% | 0.0% | 0.000 | 1.248 | 0.368 | 5.34 | |
| pSPB130 | Average (71) | 0.044 | 0.044 | 0.000 | 45.6% | 11.2% | |||||
| Best line | 0.152 | 0.015 | 0.000 | 91.2% | 39.5% | 1.320 | 0.448 | 0.120 | |||
| WKS116 | Control | 0.000 | 0.017 | 0.000 | 0.0% | 0.0% | 0.000 | 1.725 | 0.636 | 5.29 | |
| pSPB130 | Average (40) | 0.040 | 0.012 | 0.000 | 73.2% | 8.7% | |||||
| Best line | 0.051 | 0.003 | 0.000 | 94.9% | 39.3% | 1.226 | 0.245 | 0.096 | |||
| WKS124 | Control | 0.000 | 0.022 | 0.045 | 0.0% | 0.0% | 0.000 | 0.192 | 2.011 | 4.85 | |
| pSPB130 | Average (16) | 0.728 | 0.098 | 0.021 | 82.3 % | 24.6% | |||||
| Best line | 1.017 | 0.058 | 0.000 | 94.6% | 44.2% | 1.161 | 0.140 | 0.262 | |||
| WKS140 | Control | 0.000 | 0.075 | 0.000 | 0.0% | 0.0% | 0.000 | 2.412 | 0.271 | 5.25 | |
| pSPB130 | Average (49) | 0.077 | 0.063 | 0.000 | 47.3% | 11.4% | |||||
| Best line | 0.114 | 0.007 | 0.000 | 94.1% | 33.6% | 0.689 | 0.136 | 0.031 | |||
del, delphinidin; cya, cyanidin; pel, pelargonidin; del (%), delphinidin/total anthocyanidin; acyl (%), acylated anthocyanin/total anthocyanin; M, myricetin; Q, quercetin; K, kaempferol; best line, a line producing the highest percentage of delphinidin and exhibiting clear color change.
a mg g−1 fresh petals.
Only pelargonidin and cyanidin were detected as anthocyanidins, and kaempferol and quercetin as flavonols, which is consistent with previous reports (Biolley and Jay 1993, Mikanagi et al. 2000). The absence of delphinidin and myricetin confirmed the deficiency of F3′5′H activity in the petals of these cultivars. The absence of flavones, which are stronger co-pigments, was also confirmed. The cultivars WKS77, WKS82, WKS100, WKS116, WKS124 and WKS140 were selected for genetic transformation.
Cloning of flavonoid biosynthetic genes and construction of binary vectors
The aromatic acylation of anthocyanin shifts its color toward blue by 3–4 nm (Fujiwara et al. 1998). For co-expression with the F3′5′H gene in rose, the cDNA of hydroxycinnamoyl CoA:anthocyanin 5-hydroxycinnamoyl transferase (5AT) was obtained from the Torenia hybrida petal cDNA library. Torenia 5AT exhibited 37 and 35% amino acid sequence identity to gentian 5AT (Fujiwara et al. 1998) and perilla 3AT (Yonekura-Sakakibara et al. 2000), respectively. Its function was confirmed by expressing the cDNA in Escherichia coli and yeast as previously described (Fujiwara et al. 1998, Yonekura-Sakakibara et al. 2000). The enzyme catalyzed the transfer of the coumaroyl or caffeoyl moiety to 5-glucose of anthocyanidin 3,5-diglucosides (data not shown).
The substrate specificity of the DFR often determines which anthocyanidins a plant accumulates (Forkmann and Heller 1999). The DFR of some species, such as iris and gentian, which mainly accumulate delphinidin and lack pelargonidin, is expected to have a similar substrate to petunia DFR, which efficiently reduces dihydromyricetin. Iris DFR cDNA was isolated from the Iris×hollandica petal cDNA library. The iris DFR exhibited reasonable amino acid sequence identity to the DFRs of other plants (61, 58 and 56% to cymbidium, rose and petunia DFR, respectively) and had a specific feature, namely that it did not utilize dihydrokaempferol as a substrate (Johnson et al. 1999). Such substrate specificity is suitable for the conversion of the metabolic flux toward delphinidin biosynthesis when it is co-expressed with the F3′5′H gene.
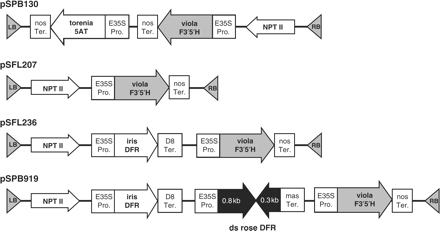
The binary vectors used in this study (pSPB130, pSFL207, pSFL236 and pSPB919) are shown in Fig. 2. The vector pSPB130 is designed for the constitutive overexpression of the viola F3′5′H BP40 gene and the torenia 5AT gene in rose. The binary vector pSFL207 is used for the constitutive overexpression of the viola F3′5′H gene alone, and pSFL236 is for the co-expression of the viola F3′5′H and the iris DFR genes. The vector pSPB919 is to down-regulate the endogenous rose DFR gene using RNA interference (RNAi) and to overexpress the iris DFR and the viola F3′5′H genes.
Schematic representation of binary vectors constructed for color modification. Only some T-DNA regions are shown. The directions of the cDNA sense strand are shown by arrows. All of them have the nptII gene as the selectable marker for plant transformation. E35S Pro., enhanced CaMV 35S promoter; mas Ter., terminator region from manopine synthase; nos Ter., nopaline synthase gene terminator; D8 Ter., terminator region from a petunia phospholipid transfer protein gene (D8) (Holton 1996); F3′5′H, flavonoid 3′,5′-hydroxylase; DFR, dihydroflavonol 4-reductase; 5AT, anthocyanin 5-acyltransferase.
Generation of transgenic roses and their phenotype
Embryogenic calli were induced from the selected cultivars and subjected to transformation with Agrobacterium tumefaciens containing pSPB130 to assess how efficiently these cultivars can accumulate delphinidin and the effect of delphinidin on flower color. At the same time, the effect of 5-acylation catalyzed by torenia 5AT on flower color could be evaluated. Transgenic roses were obtained, and flowers with altered color were subjected to flavonoid analysis. The results are summarized in Table 2. The production of delphinidin and myricetin indicates that the introduced F3′5′H gene functioned in transgenic roses. The delphinidin contents (percentage of the total anthocyanidins and amount) and the changes in flower color varied with the transgenic hosts and events.
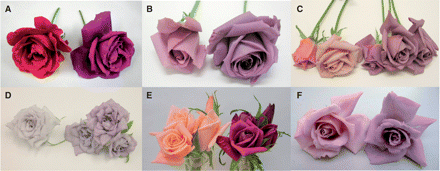
The accumulation of delphinidin conferred flower color changes. Some of them, such as WKS82, WKS100, WKS116 and WKS140 transformants (Fig. 3B, C, D, F), exhibited a novel blue hue that has not been achieved in hybridization breeding. First, WKS82/130-4-1 and WKS82/130-9-1 were selected from these transgenic roses on the basis of flower color and performance in a greenhouse, and were subjected to a field trial. Transgenic WKS77 and WKS124 also changed their flower color with delphinidin production. The amount of anthocyanins increased, and the flower color became darker. The color of the flowers remained magenta, probably because the pH of the petals was lower than that of other cultivars.
Flower color changes by delphinidin production. The rose cultivars WKS77, WKS82, WKS100, WKS116, WKS124 and WKS140 were transformed with pSPB130, and their flower color changed (left, host; right, a transformant). A flower of the line exhibiting the most significant color change is shown. (A) WKS77, (B) WKS82, (C) WKS100, (D) WKS116, (E) WKS124, (F) WKS140.
These results indicated that delphinidin production confers a blue hue to the flowers. They also revealed that both the delphinidin contents and the resultant color depend on the host cultivars. A dominant production of delphinidin is not always easy to achieve by overexpression of the F3′5′H gene even in the selected cultivars.
Only some anthocyanins (up to 44%) were revealed to be aromatically acylated by the introduced torenia 5AT gene. The effects of anthocyanin acylation on flower color were not visually observable when the anthocyanins were partly acylated because the 5-acylation of anthocyanin shifts only 4 nm to a longer wavelength (Fujiwara et al. 1998).
Transgenic plants can be used as breeding material, since the transgene is expected to be heritable by progeny. Dominant production of delphinidin irrespective of the cultivars is a desirable characteristic to incorporate into rose breeding. However, since the delphinidin content depends on cultivars to a large extent, the incorporation of the overexpression of the viola F3′5′H gene alone would not generate roses containing exclusively delphinidin. This prompted us to design another binary vector (pSPB919) in order to down-regulate an endogenous pathway that potentially competes against the introduced F3′5′H gene.
Exclusive delphinidin accumulation by functional replacement of rose DFR with iris DFR in vivo
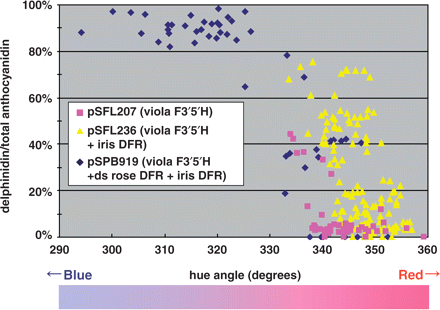
A commercial mauve cultivar, ‘Lavande’, was used to evaluate the efficacy of the binary vectors, pSFL207, pSFL236 and pSPB919 (Fig. 2), because Lavande exhibits the highest transformation frequency and the earliest flowering. The results of the transformation and anthocyanidin and flower color of the resultant transgenic flowers are summarized in Table 3 and Fig. 4. The transgenic rose flowers exhibited various degrees of flower color change, and the relationship between the hue and the delphinidin content (%) derived from pSFL207, pSFL236 and pSPB919 is summarized in Fig. 4. A higher delphinidin content yielded a bluer flower color, as shown in Fig. 4. Despite the fact the overexpression of the viola F3′5′H gene (pSFL207) successfully yielded up to 44% of delphinidin in the petals, the amount of cyanidin still surpassed that of delphinidin, about two-thirds of the transgenic plants produced <20% delphinidin and the change in flower color was not clear. Additional expression of the iris DFR gene (pSFL236) increased the delphinidin contents (Table 3, Fig. 4), indicating that the iris DFR gene was useful to change the metabolic flux toward delphinidin. The variability of the delphinidin content is large for pSFL236 plants and ranges from 0 to 75% (Fig. 4).
Correlation of delphinidin content and petal colors in transgenic Lavande. The percentage of delphinidin in the petals of individual transgenic plants was plotted against the flower color represented by the hue value in degrees (hue angle). Pure red and blue have hue values of 360 and 270° in the hue angle, respectively. The higher the percentage of delphinidin was, the bluer the flower color became. The color gradation bar approximately indicates the corresponding petal color of transgenic Lavande petals.
Summary of the flavonoid composition in transgenic Lavande
| Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | Flavonol (mg g−1)a . | |||
|---|---|---|---|---|---|---|---|
| . | . | del . | cya . | . | M . | Q . | K . |
| Control | 0.000 | 0.109 | 0.0% | 0.000 | 1.020 | 0.195 | |
| pSFL207 | Average (75) | 0.009 | 0.106 | 9.3% | |||
| Best line | 0.052 | 0.066 | 44.2% | 0.216 | 0.312 | 0.010 | |
| pSFL236 | Average (145) | 0.048 | 0.150 | 24.1% | |||
| Best line | 0.141 | 0.046 | 75.3% | 0.159 | 0.387 | 0.013 | |
| pSPB919 | Average (48) | 0.045 | 0.019 | 78.7% | |||
| Best line | 0.101 | 0.002 | 98.2% | 0.420 | 0.135 | 0.008 | |
| Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | Flavonol (mg g−1)a . | |||
|---|---|---|---|---|---|---|---|
| . | . | del . | cya . | . | M . | Q . | K . |
| Control | 0.000 | 0.109 | 0.0% | 0.000 | 1.020 | 0.195 | |
| pSFL207 | Average (75) | 0.009 | 0.106 | 9.3% | |||
| Best line | 0.052 | 0.066 | 44.2% | 0.216 | 0.312 | 0.010 | |
| pSFL236 | Average (145) | 0.048 | 0.150 | 24.1% | |||
| Best line | 0.141 | 0.046 | 75.3% | 0.159 | 0.387 | 0.013 | |
| pSPB919 | Average (48) | 0.045 | 0.019 | 78.7% | |||
| Best line | 0.101 | 0.002 | 98.2% | 0.420 | 0.135 | 0.008 | |
del, delphinidin; cya, cyanidin; del (%), delphinidin/total anthocyanidin; M, myricetin; Q, quercetin; K, kaempferol; best line, a line producing the highest percentage of delphinidin.
a mg g−1 fresh petals.
Summary of the flavonoid composition in transgenic Lavande
| Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | Flavonol (mg g−1)a . | |||
|---|---|---|---|---|---|---|---|
| . | . | del . | cya . | . | M . | Q . | K . |
| Control | 0.000 | 0.109 | 0.0% | 0.000 | 1.020 | 0.195 | |
| pSFL207 | Average (75) | 0.009 | 0.106 | 9.3% | |||
| Best line | 0.052 | 0.066 | 44.2% | 0.216 | 0.312 | 0.010 | |
| pSFL236 | Average (145) | 0.048 | 0.150 | 24.1% | |||
| Best line | 0.141 | 0.046 | 75.3% | 0.159 | 0.387 | 0.013 | |
| pSPB919 | Average (48) | 0.045 | 0.019 | 78.7% | |||
| Best line | 0.101 | 0.002 | 98.2% | 0.420 | 0.135 | 0.008 | |
| Vector . | . | Anthocyanidin (mg g−1)a . | del (%) . | Flavonol (mg g−1)a . | |||
|---|---|---|---|---|---|---|---|
| . | . | del . | cya . | . | M . | Q . | K . |
| Control | 0.000 | 0.109 | 0.0% | 0.000 | 1.020 | 0.195 | |
| pSFL207 | Average (75) | 0.009 | 0.106 | 9.3% | |||
| Best line | 0.052 | 0.066 | 44.2% | 0.216 | 0.312 | 0.010 | |
| pSFL236 | Average (145) | 0.048 | 0.150 | 24.1% | |||
| Best line | 0.141 | 0.046 | 75.3% | 0.159 | 0.387 | 0.013 | |
| pSPB919 | Average (48) | 0.045 | 0.019 | 78.7% | |||
| Best line | 0.101 | 0.002 | 98.2% | 0.420 | 0.135 | 0.008 | |
del, delphinidin; cya, cyanidin; del (%), delphinidin/total anthocyanidin; M, myricetin; Q, quercetin; K, kaempferol; best line, a line producing the highest percentage of delphinidin.
a mg g−1 fresh petals.
On the other hand, more than two-thirds of the transgenic rose plants harboring pSPB919 produced >80% delphinidin. These lines contained exclusively delphinidin, which indicates that the successful conversion of cyanidin synthesis to delphinidin synthesis was achieved (Table 3). Some of them (LA/919-4-10, etc.) exhibited the bluest hue (Fig. 5), and the Royal Horticultural Society Color Chart (RHSCC) number of their petals was 85b (Violet group), while that of the control Lavande flower was 186c (Greyed-purple group). It is noteworthy that the transgenic plants harboring pSPB919 exhibited a higher delphinidin content and showed much less variation in delphinidin content than those harboring pSFL207 and pSFL236. The results indicate that the rose DFR gene was down-regulated and the iris DFR gene was expressed. In other words, the rose DFR was functionally replaced with the iris DFR in vivo. Myricetin was detected in the transgenic plants (Table 3), which confirmed that the introduced F3′5′H gene functioned.
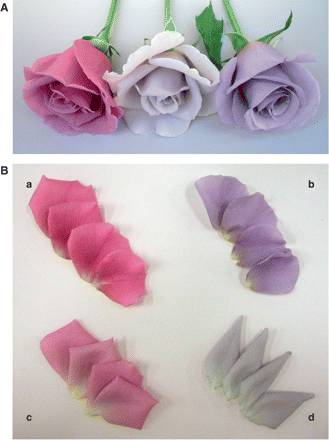
Flower and petal color comparison. The pink-flowered rose cultivar Lavande (A, left and B, a) was transformed with pSPB919. The resultant transgenic plants including LA/919-4-10 produced violet-colored transgenic flowers (A, right and B, b) containing 98% delphinidin. The flower color is evidently bluer than that of the hosts, the commercial varieties Madam Violet (B, c) and Seiryu (B, d). Madam Violet and Seiryu are regarded as the bluest of the current rose varieties. Some of the transgenic roses derived from pSPB919 exhibited paler flower color and contained decreased amounts of anthocyanidin (A, center).
Some of the transgenic roses derived from pSPB919 exhibited a paler flower color and contained less anthocyanidin (Fig. 5A, center). This is probably due to suppression of the endogenous DFR gene and inefficient complementation by the introduced iris DFR gene.
Analysis of anthocyanins
The anthocyanins in LA/919-4-10 petals were analyzed in detail. The ratio of anthocyanin in petals is 90.0% delphinidin 3,5-diglucoside and 10.0% delphinidin 3-glucoside. The identities of these pigments were confirmed with co-chromatography with authentic compounds using HPLC (data not shown). Their structures were further confirmed by time-of flight-mass spectometry (TOF-MS) and nuclear magnetic resonance (NMR). The host Lavande accumulates 96.0% cyanidin 3,5-diglucoside and 4.0% cyanidin 3-glucoside. The presence of 3-monoglucoside and 3,5-diglucoside in R. hybrida and wild roses has been previously reported (Biolley and Jay 1993, Mikanagi et al. 2000), although it was recently proposed that the 5-glucosylation of anthocyanins proceeds to 3-glucosylation in R. hybrida (Ogata et al. 2005).
Molecular analysis of the transgenic rose LA/919-4-10
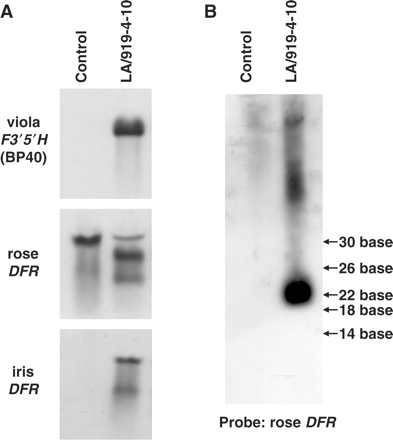
The results of the Northern analysis of LA/919-4-10 are shown in Fig. 6A. The transcripts of the viola F3′5′H and iris DFR genes were observed to have intact sizes, while only smaller size bands of the rose DFR mRNA were detected. Small interfering RNA (siRNA) of about 23 bp was detected when using the rose DFR cDNA as the probe. These results reveal that the rose endogenous DFR mRNA was successfully degraded by the introduced RNAi cassette of the rose DFR (Fig. 6B). Southern analysis of LA/919-4-10 with the viola F3′5′H, iris DFR, rose DFR and NPTII genes indicates that the plant contains single-copy T-DNA (data not shown), which prompted us to breed this rose as the parent.
Northern blot analysis of LA/919-4-10. The expected sizes of the transcripts of viola F3′5′H BP40 (1.8 kb) and iris DFR (1.7 kb) genes were observed, while only the smaller size was detected for rose DFR mRNA (A). A rose DFR probe detected about 23 bp of the small sized RNA, which was supposed to be a degraded endogenous rose DFR transcript with RNAi (B).
Hybridization breeding
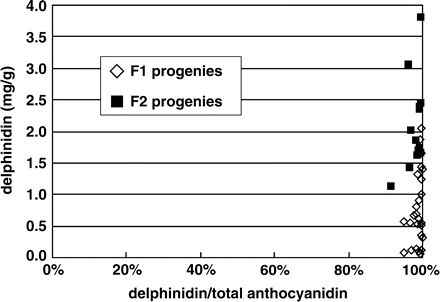
In order to evaluate the efficacy of the redirection of the pathway from cyanidin to delphinidin, the pollen of LA/919-4-10 was hybridized with a seed parent ‘Black Baccara’, a dark-red variety that accumulates a large amount of cyanidin (Table 1). About 50% of the seeds germinated normally in the kanamycin media, and delphinidin was detected in all of the kanamycin-resistant F1 progeny. The delphinidin content was 99.9% at maximum (average: 98.7%) in these progeny. Some delphinidin-producing F1 plants were backcrossed to Black Baccara. The anthocyanidin analysis of the F2 progeny revealed that the flowers also contained delphinidin almost exclusively (Fig. 7). The results indicate that the character of exclusive accumulation of delphinidin is heritable as a single locus. Although the flower color varied from red-purple to deep magenta depending on the genetic background, probably their pH and the amount of flavonols, the introduced transgenes were able to convert the metabolic flux completely toward delphinidin. Complete modification of the metabolic flux toward delphinidin should confer the ability to produce delphinidin exclusively in the progeny.
Delphinidin contents of the transgenic progeny. The accumulation of delphinidin was confirmed in all of the KmR progeny of LA/919-4-10. The flowers of the F1 and F2 progeny contained exclusively delphinidin.
Discussion
In this study, we selected rose cultivars that are suitable for delphinidin production and color change toward blue. Constitutive expression of the viola F3′5′H gene successfully resulted in delphinidin accumulation and color changes with a blue hue that has commercial value as far as novel flower color is concerned. More efficient and dominant modification of the flavonoid pathway toward delphinidin was achieved by functional replacement of DFRs, i.e. the down-regulation of the endogenous DFR gene and overexpression of the iris DFR gene in vivo, in addition to the overexpression of the viola F3′5′H gene. The ability for the exclusive accumulation of delphinidin was heritable by progeny, indicating that the flavonoid pathway was consistently modified toward delphinidin production.
Overexpression of the F3′5′H gene in selected cultivars
The biggest advantage of genetic engineering in plant breeding is that a novel gene from a heterologous source can be utilized to improve a plant, which liberates plant breeding from genetic constraints. However, the overexpression of a heterologous gene alone does not always efficiently yield a desired compound, as reported above. In order to achieve the accumulation of a desirable amount of a compound, it is necessary to select host cultivars that have the least competing reaction against an introduced gene, as in the case of the blue carnation (Holton 1996). In the case of color modification toward blue, higher amounts of flavonols and higher vacuolar pH are also desirable. Even when delphinidin accumulated in the selected cultivars, the flower color varied depending on the cultivars (Table 2, Fig. 3). Since cultivars such as WKS82, WKS100, WKS116 and WKS140, which give a blue hue, contain large amounts of flavonols and have a higher pH, flavonols may contribute to the blue coloration by co-pigmentation.
The delphinidin content (%) and flower color varied depending on the cultivar, and, even in selected cultivars, it depended on the transgenic events, as shown in Table 2 and Fig. 4 (pSFL207 and pSFL236 plants). Such variations of phenotypes have often been observed in transgenic plants (Filipecki and Malepszy 2006). The generation of many independent transgenic plants in various host cultivars and selection of lines having desirable phenotypes among them is a practical procedure to obtain transgenic plants that have commercial value.
Functional replacement of the rose DFR with the iris DFR resulted in the cultivar-independent exclusive accumulation of delphinidin
The exclusive production of delphinidin and the most significant color change toward blue were achieved with pSPB919 in Lavande. We succeeded in the replacement of the function of rose DFR genes with an iris DFR gene in transgenic roses by using RNAi. We previously showed that RNAi gave more efficient gene suppression than antisense or sense suppression (Nakamura et al. 2006). The degradation of rose DFR transcripts and overexpression of the iris DFR were confirmed by Northern analysis. The replacement was efficient; a higher frequency and less deviated delphinidin accumulation and color change were observed.
Moreover, the flowers of the F1 and F2 progeny of LA/919-4-10 exclusively contained delphinidin. Such a dominant character conferred by the transgenes is very useful for the incorporation of a novel ability (delphinidin production) in rose breeding, which has suffered from the limited ability to synthesize only pelargonidin and cyanidin as anthocyanidins. The ability to synthesize delphinidin achieved in this study will change rose breeding and increase rose varieties as far as flower color is concerned. The fact that the flower color of the progeny is not as blue as that of LA/919-4-10 indicates that other factors, such as the presence of a suitable amount of flavonols (co-pigments) and a higher vacuolar pH, are necessary to achieve blue color in addition to delphinidin production. Since hybridization breeding can generate host rose cultivars with various genetic backgrounds, delphinidin production in these backgrounds will yield various novel color tones, in addition to blue, in their flowers.
Replacement of the enzymatic function achieved in this study should be useful to engineer various plant metabolic pathways not limited to the flavonoid or other secondary metabolic pathways. Although replacement of an endogenous gene by homologous recombination would be desirable, homologous recombination in plants has only been reported in a few model plants (Terada et al. 2007). Many commercially important plants have high ploidy. For example, cultivated roses and chrysanthemums are tetraploids and hexaploids. The functional replacement depending on post-transcriptional gene silencing shown in this study is practical in plants having high ploidy.
Toward bluer/future perspectives
The production of delphinidin-based anthocyanins in heterologous plants has been the preferable strategy to generate flowers with a blue hue, which is also achieved in rose in this study. In addition to delphinidin production, blue flowers generate their flower color through the combination of various strategies, such as highly modified anthocyanins, a stable complex with co-pigments, a higher vacuolar pH and metal ions, as reported above. The reconstitution of these factors in roses should generate bluer color. However, this is a very challenging task. One reason is that many genes have to be properly expressed in roses, and the genes involved in vacuolar pH regulation and metal ion uptake have not been well characterized.
Therefore, a feasible strategy toward bluer follows. The co-existence of delphinidin-based anthocyanins and a flavone compound gave bluer color in a transgenic carnation (Fukui et al. 2003) than delphinidin alone. In this case, the flavone was derived from a host carnation cultivar. The co-expression of a flavone synthase gene (Akashi et al. 1999) in roses may result in a bluer color than that obtained in this study. The multiple aromatic acylation of anthocyanins generates a stable blue color (Honda and Saito 2002). We demonstrated that the rose can synthesize aromatically acylated anthocyanins by the expression of torenia 5AT. Gentian 5,3′AT (Mizutani et al. 2006) and Clitoria ternatea AT (Noda et al. 2006), which can add multiple acyl moieties to anthocyanins, are useful molecular tools. The efficient acylation of anthocyanins may not be easy, since the acylation by torenia 5AT remained at <40%.
Although more effort will be required to achieve sky-blue roses, the exclusive delphinidin production and the resultant color changes presented in this study represent a historic milestone in rose breeding.
Materials and Methods
Plant materials and plant tissue culture
Flowers of rose cultivars were screened and harvested in Keisei Rose Nurseries (Sawara, Japan) in 1997–2000. Stem internodes were harvested from rose plants grown in a greenhouse, and the surface was sterilized by immersing them in a 5% (v/v) sodium hypochlorite solution for 5 min and then rinsing them with sterilized water. The explants (1 cm in length) were prepared from the stem internodes and cultured at 25°C under 16 h/8 h conditions in a medium consisting of Murashige and Skoog mineral salts and vitamins, 30 g l−1 sucrose, 12 g l−1 agar and 1 mg l−1 6-benzylaminopurine (BA). Rose transformation was described previously (Firoozabady et al. 1994) in a study using Agrobacterium tumefaciens Agl0 (Lazo et al. 1991). The acclimated transgenic plants were potted and then grown in contained greenhouses until blooming.
Molecular procedures
Most molecular procedures, including cDNA library construction, screening of the library, binary vector construction and Southern/Northern analysis, were described previously (Fukuchi-Mizutani 2003). The detection of siRNA was carried out as described previously using digoxogenin-labeled rose DFR cDNA as a probe, as previously described (Goto et al. 2003).
The procedures involving binary vectors for rose transformation were also described previously (Fukuchi-Mizutani et al. 2003). In brief, pBin19 and pBinPLUS (van Engelen et al. 1995), which contain neomycin phosphotransferase II (NPTII) as the selection marker, were used to construct a binary vector, and the enhanced cauliflower mosaic virus (CaMV) 35S promoter derived from pE2113 (Mitsuhara et al. 1996) was used to regulate the transgenes. Torenia anthocyanin 5-acyltransferase cDNA (database accession No. AB332099) was obtained from its petal cDNA library (Suzuki 2000) as described previously (Yonekura-Sakakibara et al. 2000). Iris×hollandica DFR cDNA (AB332098) was cloned from its petal cDNA (Imayama et al. 2004) as described before (Tanaka et al. 1995). Viola spp. contains two F3′5′H genes (Brugliera et al. 2004), and one of them (clone BP40) was used in this study (AB332097).
Measurement of flower color and petal pH
The flower petal color was evaluated using colorimetric values in a CIE L*a*b* system measured with a CM-2022 spectrophotometer (Minolta Co., Ltd, Tokyo, Japan), and the data were calculated under a 10° observer and D65 illuminants as described before (Fukui et al. 2003). Color data were analyzed using SpectraMagic™ color-control software (Minolta). The RHSCC number which was closest to the flower color was selected by Color Classification System version 2.1.1 (The Japan Research Institute Co., Ltd; Shibano et al. 2002), which is based on the color value (CIE L*a*b* color system) obtained by visual discrimination and measurement with the device mentioned above. This system can be used for objective selection of the closest RHSCC number by color data. RHSCC is a standard reference for plant color identification in the horticultural industry.
Fresh rose petals (2 g) were frozen at −80°C and homogenized. The pH of the pressed juice was measured with an F-22 pH meter with a 6069-10C electrode (Horiba Ltd, Kyoto, Japan).
Analysis of anthocyanins
Rose anthocyanins were extracted with 50% (v/v) acetonitrile containing trifluoroacetic acid (TFA) as previously reported (Fukui et al. 2003). HPLC was conducted using an RSpak DE-413L (25 cm × 4.6 mm, Shoko Co., Ltd, Tokyo, Japan) column and a flow rate of solvent of 0.6 ml min−1; the solvent system used was as follows: after a 15 min linear gradient elution from 10 to 50% of acetonitrile containing 0.5% TFA in H2O, 10 min of isocratic elution of 50% of acetonitrile containing 0.5% TFA in H2O was carried out, and detection was conducted with a photodiode array detector SPD-M10A (Shimadzu Co., Ltd, Kyoto, Japan) that can measure absorbance at 600–250 nm. The retention times of aromatically acylated anthocyanins were around 19 min in this analysis. The anthocyanin spectra were measured with a photodiode array. Aromatically acylated anthocyanins can be identified because they exhibit a longer wave shift and a peak at 280 nm. TOF-MS and NMR analyses were described before (Fukui et al. 2003).
Analysis of anthocyanidins
Petal extracts prepared as described previously (Fukui et al. 2003) were dissolved in 6 N HCl (0.2 ml) and kept at 100°C for 20 min. The hydrolyzed anthocyanidins were extracted with 0.2 ml of 1-pentanol. HPLC was performed using an ODS-A312 (15 cm × 6 mm, YMC Co., Ltd, Kyoto, Japan) column, a flow rate of solvent of 1 ml min−1, and detection at an absorbance of 600–400 nm on a SPD-M10A photodiode array detector (Shimadzu Co., Ltd). The solvent system used was as follows: acetic acid : methanol : water= 15 : 20 : 65. Under these HPLC conditions, the retention time and λmax of delphinidin were 4.0 min and 534 nm, respectively, and these values were compared with those of authentic delphinidin chloride (Funakoshi Co., Ltd, Tokyo, Japan).
Analysis of flavonols
Rose hosts and some of the transgenic rose flowers were subjected to flavonol analysis. Petal extracts were hydrolyzed in 0.2 ml of 0.1 M potassium phosphate buffer (pH 4.5) with 6 U of β-glucosidase (Shin-nihon Chemical Co., Ltd, Japan) and 1 U of naringinase (Sigma Chemical Co., St Louis MO, USA) under incubation for 16 h at 30°C. Acetonitrile (0.2 ml, 90%) was added to the reaction mixture, which was then subjected to HPLC analysis. HPLC was conducted using a Develosil C30-UG-5 (15 cm × 4.6 mm, YMC Co., Ltd) column and a flow rate of solvent of 0.6 ml min−1; the solvent system used was as follows: after a 10 min linear gradient elution using 18–63% acetonitrile containing 0.1% TFA in H2O, 5 min of isocratic elution of 63% acetonitrile containing 0.1% TFA in H2O was carried out with absorbance at 400–250 nm detected using the photodiode array detector SPD-M10A (Shimadzu Co., Ltd).
Rose hybridization
Hybridization breeding was carried out using a transformant (LA/919-4-10) as a pollen parent and a dark-red rose variety, ‘Black Baccara’ (Meilland International), as the seed parent. Fruits were collected on the 100th day after pollination. Seed germination was accomplished by peeling the achenes and embedding them in moistened filter paper in plastic dishes. The water used for seed germination was sterilized water containing 1.0 ml l−1 PPM™ (Plant Preservative Mixture; Plant Cell Technology, Inc., Washington, DC, USA) and 50 mg l−1 kanamycin, and seedlings were raised by potting only normally budded plants with green cotyledons.
Acknowledgments
The authors are grateful to Keisei Rose Nurseries, Inc., for their long and dedicated provision of plant materials for the hosts of genetic transformation and their knowledge and expertise on roses. Dr. Yuko Ohashi (National Institute of Agrobiological Sciences) and Dr. Robert Ludwig (University of California, Santa Cruz) are acknowledged for providing the enhanced promoter and A. tumefaciens Agl0, respectively. We also thank Dr. Akira Kanazawa (Hokkaido University) for his advice on RNAi analysis and all those who collaborated on the blue rose project. The authors gratefully acknowledge the invaluable contribution to this project by Professor Edwina Cornish (Monash University) and Dr. Takaharu Tanaka (Suntory Limited), and Dr. Yasuyuki Yamada (Professor Emeritus of Kyoto University) for his continuous support and encouragement. We appreciate Dr. Stephen F. Chandler (Florigene Pty. Ltd) for his critical reading of the manuscript, and our colleagues and technical staff for their many years of effort.
References
Abbreviations:
-
AT
hydroxycinnamoyl CoA:anthocyanin hydroxycinnamoyl transferase
-
CaMV
cauliflower mosaic virus
-
DFR
dihydroflavonol 4-reductase
-
F3′5′H
flavonoid 3′,5′-hydroxylase
-
NMR
nuclear magnetic resonance
-
NPTII
neomycin phosphotransferase
-
RHSCC
Royal Horticultural Society Color Chart
-
RNAi
RNA interference
-
siRNA
small interfering RNA
-
TFA
trifluoroacetic acid
-
TOF-MS
time-of-flight-mass spectrometry.
Author notes
Present address: RIKEN Plant Science Center, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama City, Kanagawa, 230-0045 Japan.