-
PDF
- Split View
-
Views
-
Cite
Cite
Xiaoyun Dong, Edward L. Braun, Erich Grotewold, Functional Conservation of Plant Secondary Metabolic Enzymes Revealed by Complementation of Arabidopsis Flavonoid Mutants with Maize Genes, Plant Physiology, Volume 127, Issue 1, September 2001, Pages 46–57, https://doi.org/10.1104/pp.127.1.46
Close - Share Icon Share
Abstract
Mutations in the transparent testa(tt) loci abolish pigment production in Arabidopsis seed coats. The TT4, TT5, andTT3 loci encode chalcone synthase, chalcone isomerase, and dihydroflavonol 4-reductase, respectively, which are essential for anthocyanin accumulation and may form a macromolecular complex. Here, we show that the products of the maize (Zea mays)C2, CHI1, and A1 genes complement Arabidopsis tt4, tt5, andtt3 mutants, restoring the ability of these mutants to accumulate pigments in seed coats and seedlings. Overexpression of the maize genes in wild-type Arabidopsis seedlings does not result in increased anthocyanin accumulation, suggesting that the steps catalyzed by these enzymes are not rate limiting in the conditions assayed. The expression of the maize A1 gene in the flavonoid 3′ hydroxylase Arabidopsis tt7 mutant resulted in an increased accumulation of pelargonidin. We conclude that enzymes involved in secondary metabolism can be functionally exchangeable between plants separated by large evolutionary distances. This is in sharp contrast to the notion that the more relaxed selective constrains to which secondary metabolic pathways are subjected is responsible for the rapid divergence of the corresponding enzymes.
A general characteristic of plants is their ability to accumulate a large number of secondary compounds. Many of these secondary metabolites play unique roles in the interaction between plants and the environment, therefore increasing evolutionary fitness. Secondary metabolism provides unique opportunities to investigate the evolution of complex metabolic networks in the absence of the tight selective constrains characteristic of primary metabolism (Pichersky and Gang, 2000). Some secondary metabolic pathways are unique to particular groups of plants, whereas others are more broadly distributed.
Flavonoids are 15-carbon phenolic compounds synthesized by many different plants. Flavonoid biosynthesis is one of the best described plant secondary metabolic pathways, and genes encoding flavonoid biosynthetic enzymes have been cloned and characterized in various species. The isolation of flavonoid biosynthetic genes has been aided by the characterization of mutations in model systems such as maize (Zea mays), petunia (Petunia hybrida), snapdragon (Antirrhinum majus), and, more recently, Arabidopsis (Dooner et al., 1991; Shirley, 1996; Mol et al., 1998).
Products synthesized by the early steps of the flavonoid pathway are found in bryophytes and ferns, whereas gymnosperms and angiosperms accumulate additional classes of flavonoids, probably reflecting the recruitment of more genes to flavonoid biosynthesis, as well as the evolution of novel functions played by these compounds. In angiosperms, flavonoids function to protect plants from predators and infectious agents, to shield plants from UV-B radiation, as signal molecules in plant-bacterium symbioses, and as pigments to attract pollinators and seed dispersers (Koes et al., 1994). Despite the wide distribution of this large group of compounds among the flowering plants, particular classes of flavonoids have distinct functions in the different plant groups. For example, flavonols are essential for male fertility in maize, petunia, and tobacco (Nicotiana tabacum; Mo et al., 1992; Ylstra et al., 1992), but not in Arabidopsis (Burbulis et al., 1996; Ylstra et al., 1996). Isoflavonoids are the major phytoalexins in legumes (Dixon and Steele, 1999), whereas similar roles are played by 3-deoxyanthocyanidins in sorghum Sorghum bicolor) and other grasses (Snyder and Nicholson, 1990). The flexibility of this and other secondary metabolic pathways suggests that the selective pressures upon genes encoding enzymes involved in secondary metabolism are quite variable. This fact, coupled with the observation that some enzymes involved in secondary metabolism have arisen from independent ancestors (Pichersky and Gang, 2000), makes it unclear whether mutations in these genes can be complemented by genes from distantly related organisms.
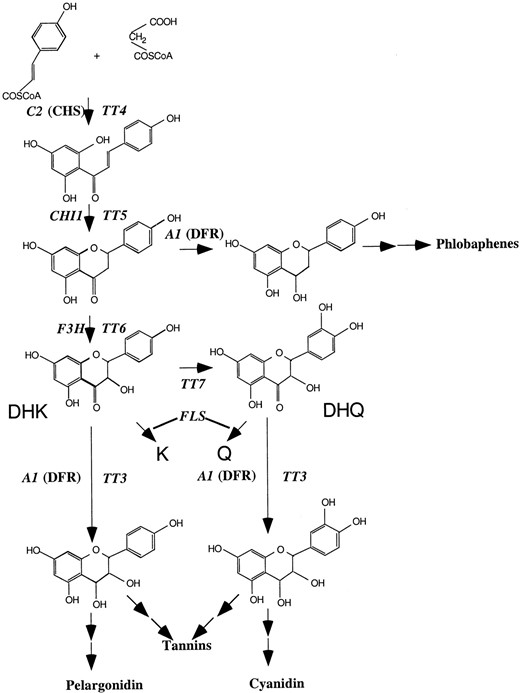
Flavonoid biosynthetic pathway. Only the enzymatic steps significant for the studies presented here are indicated, with the Arabidopsis and maize genes labeled. Dihydrokaempferol (DHK), kaempferol (K), dihydroquercetin (DHQ), quercetin (Q).
In Arabidopsis, single genes encode many flavonoid biosynthetic enzymes including CHS and DFR. Mutations that completely abolish flavonoid or anthocyanin production have been identified in genes encodingCHS, CHI, and DFR (Shirley et al., 1995; Shirley, 1996; Pelletier et al., 1999). This is in sharp contrast with other plants, where small multigene families encode many of the biosynthetic enzymes of the pathway (Dooner et al., 1991). The Arabidopsis CHS protein is more than 85% identical to the maize C2, whereas the Arabidopsis CHI and DFR enzymes are less than 60% identical to maize CHI1 and A1, respectively. The more limited conservation observed for CHI and DFR, coupled with the apparent absence of a requirement for CHI expression for flavonoid accumulation in maize cells (Grotewold et al., 1998), raises the fundamental question of whether enzymes present in these distantly related groups of flowering plants are exchangeable.
To explore the evolutionary conservation of enzymes involved in plant metabolic pathways, we have complemented Arabidopsis tt4,tt5, and tt6 mutants using the maize C2, CHI1, and A1 genes. The flavonoids and anthocyanins formed in complemented mutants are almost indistinguishable from those found in wild-type Arabidopsis plants. Furthermore, similar amounts of anthocyanins were found in wild-type and transgenic plants expressing the maize genes, suggesting that none of these enzymes imposes a flux constraint upon the Arabidopsis flavonoid biosynthetic pathway under the conditions tested. Only the ability of the maize A1 gene to increase the accumulation of pelargonidins in tt7 mutants (Fig. 1) suggests the possibility of different substrate specificities for maize and Arabidopsis DFR enzymes. Together, our findings provide the first experimental evidence that multiple enzymes of a secondary metabolic pathway are exchangeable between plants separated by more than 100 million years of evolution.
RESULTS
Maize C2 , CHI1, andA1 Genes Complement Pigmentation of Arabidopsistt4, tt5, and tt3 Mutants
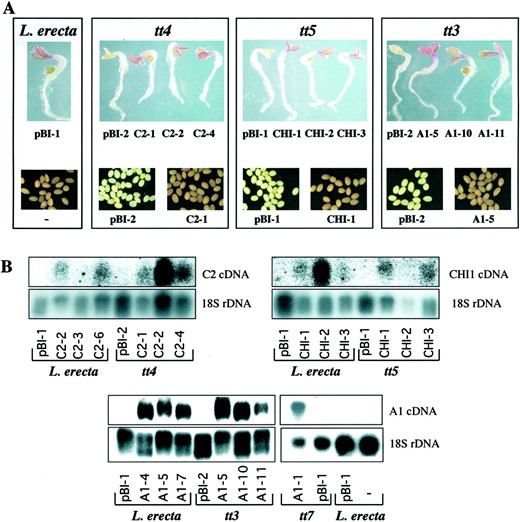
Complementation of the pigmentation of Arabidopsistt4, tt5, and tt3 mutant seedlings and seed coats with the maize C2, CHI1, andA1 genes, respectively. A, Pigment accumulation of Landsbergerecta seedlings grown in low-nitrogen media and of Landsberg erecta seed coats. Complementation of thett4 mutant (transformed with the empty pBIB121plasmid; left colorless seedling) by 35S::C2 (three independent transformation events; red seedlings); tt5 mutant (transformed with the empty pBIB121plasmid; left colorless seedling) by 35S::CHI1 (three independent transformation events; red seedlings); and tt3 mutant (transformed with the empty pBIB121 plasmid; left colorless seedling) by 35S::A1 (three independent transformation events; red seedlings). Transgenic T2 seeds are visualized under visible light. B, Northern analysis of total RNA obtained from wild-type (Landsbergerecta) and mutant (tt4, tt5, and tt3) seedlings grown in low-nitrogen media expressing C2(Landsberg erecta and tt4), CHI1(Landsberg erecta and tt5), and A1(Landsberg erecta and tt3), or from 4-week-old plants (Landsberg erecta or tt7) expressing the maize A1 gene. A probe corresponding to the 18S rRNA was used as a loading control. pBI- corresponds to lines that carry the empty pBIB121 plasmid.
The maize C2 gene, which encodes CHS (Wienand et al., 1986), was expressed from the constitutive cauliflower mosaic virus (CaMV) 35S promoter in tt4 plants (tt4-1 allele, TableI). This resulted in T1 seeds that, when germinated in low-nitrogen media, exhibited full complementation of the deficiency in anthocyanin accumulation of tt4 mutant seedlings (Fig. 2A). Seeds derived from kanamycin-resistant T2 plants showed the pigmentation characteristic of wild-type Arabidopsis (Fig. 2A,tt4). In addition, the expression of the maize C2gene resulted in the accumulation of compounds that absorb UV radiation, with the consequent disappearance of the fluorescence characteristic of tt4 seed (not shown). Furthermore, the expression of C2 in the tt4 mutants complemented the frequent germination problems that we encountered associated with thett4 mutant, but not with the tt3 ortt5 mutants (not shown).
Arabidopsis transgenic lines used in this study
| Transgene . | Line . | tt4-1 . | tt5-1 . | tt3-1 . | tt7-1 . |
|---|---|---|---|---|---|
| pBIB121 | Landsberg erecta pBI-1 | tt4 pBI-2 | tt5 pBI-1 | tt3 pBI-2 | tt7 pBI-1 |
| Landsberg erecta pBI-6 | tt4 pBI-4 | tt5 pBI-3 | tt3 pBI-4 | tt7 pBI-2 | |
| Landsberg erecta pBI-19 | tt4 pBI-5 | tt5 pBI-4 | tt3 pBI-6 | tt7 pBI-3 | |
| 35S∷C2 | Landsberg erecta C2-2 | tt4 C2-1 | – | – | – |
| Landsberg erecta C2-3 | tt4 C2-2 | – | – | – | |
| Landsberg erecta C2-6 | tt4 C2-4 | – | – | – | |
| 35S∷CHI1 | Landsberg erecta CHI-1 | – | tt5 CHI-1 | – | – |
| Landsberg erecta CHI-2 | – | tt5 CHI-2 | – | – | |
| Landsberg erecta CHI-3 | – | tt5 CHI-3 | – | – | |
| 35S∷A1 | Landsberg erecta A1-4 | – | – | tt3 A1-5 | tt7 A1-1 |
| Landsberg erecta A1-5 | – | – | tt3 A1-10 | tt7 A1-2 | |
| Landsberg erecta A1-7 | – | – | tt3 A1-11 | tt7 A1-3 |
| Transgene . | Line . | tt4-1 . | tt5-1 . | tt3-1 . | tt7-1 . |
|---|---|---|---|---|---|
| pBIB121 | Landsberg erecta pBI-1 | tt4 pBI-2 | tt5 pBI-1 | tt3 pBI-2 | tt7 pBI-1 |
| Landsberg erecta pBI-6 | tt4 pBI-4 | tt5 pBI-3 | tt3 pBI-4 | tt7 pBI-2 | |
| Landsberg erecta pBI-19 | tt4 pBI-5 | tt5 pBI-4 | tt3 pBI-6 | tt7 pBI-3 | |
| 35S∷C2 | Landsberg erecta C2-2 | tt4 C2-1 | – | – | – |
| Landsberg erecta C2-3 | tt4 C2-2 | – | – | – | |
| Landsberg erecta C2-6 | tt4 C2-4 | – | – | – | |
| 35S∷CHI1 | Landsberg erecta CHI-1 | – | tt5 CHI-1 | – | – |
| Landsberg erecta CHI-2 | – | tt5 CHI-2 | – | – | |
| Landsberg erecta CHI-3 | – | tt5 CHI-3 | – | – | |
| 35S∷A1 | Landsberg erecta A1-4 | – | – | tt3 A1-5 | tt7 A1-1 |
| Landsberg erecta A1-5 | – | – | tt3 A1-10 | tt7 A1-2 | |
| Landsberg erecta A1-7 | – | – | tt3 A1-11 | tt7 A1-3 |
Arabidopsis transgenic lines used in this study
| Transgene . | Line . | tt4-1 . | tt5-1 . | tt3-1 . | tt7-1 . |
|---|---|---|---|---|---|
| pBIB121 | Landsberg erecta pBI-1 | tt4 pBI-2 | tt5 pBI-1 | tt3 pBI-2 | tt7 pBI-1 |
| Landsberg erecta pBI-6 | tt4 pBI-4 | tt5 pBI-3 | tt3 pBI-4 | tt7 pBI-2 | |
| Landsberg erecta pBI-19 | tt4 pBI-5 | tt5 pBI-4 | tt3 pBI-6 | tt7 pBI-3 | |
| 35S∷C2 | Landsberg erecta C2-2 | tt4 C2-1 | – | – | – |
| Landsberg erecta C2-3 | tt4 C2-2 | – | – | – | |
| Landsberg erecta C2-6 | tt4 C2-4 | – | – | – | |
| 35S∷CHI1 | Landsberg erecta CHI-1 | – | tt5 CHI-1 | – | – |
| Landsberg erecta CHI-2 | – | tt5 CHI-2 | – | – | |
| Landsberg erecta CHI-3 | – | tt5 CHI-3 | – | – | |
| 35S∷A1 | Landsberg erecta A1-4 | – | – | tt3 A1-5 | tt7 A1-1 |
| Landsberg erecta A1-5 | – | – | tt3 A1-10 | tt7 A1-2 | |
| Landsberg erecta A1-7 | – | – | tt3 A1-11 | tt7 A1-3 |
| Transgene . | Line . | tt4-1 . | tt5-1 . | tt3-1 . | tt7-1 . |
|---|---|---|---|---|---|
| pBIB121 | Landsberg erecta pBI-1 | tt4 pBI-2 | tt5 pBI-1 | tt3 pBI-2 | tt7 pBI-1 |
| Landsberg erecta pBI-6 | tt4 pBI-4 | tt5 pBI-3 | tt3 pBI-4 | tt7 pBI-2 | |
| Landsberg erecta pBI-19 | tt4 pBI-5 | tt5 pBI-4 | tt3 pBI-6 | tt7 pBI-3 | |
| 35S∷C2 | Landsberg erecta C2-2 | tt4 C2-1 | – | – | – |
| Landsberg erecta C2-3 | tt4 C2-2 | – | – | – | |
| Landsberg erecta C2-6 | tt4 C2-4 | – | – | – | |
| 35S∷CHI1 | Landsberg erecta CHI-1 | – | tt5 CHI-1 | – | – |
| Landsberg erecta CHI-2 | – | tt5 CHI-2 | – | – | |
| Landsberg erecta CHI-3 | – | tt5 CHI-3 | – | – | |
| 35S∷A1 | Landsberg erecta A1-4 | – | – | tt3 A1-5 | tt7 A1-1 |
| Landsberg erecta A1-5 | – | – | tt3 A1-10 | tt7 A1-2 | |
| Landsberg erecta A1-7 | – | – | tt3 A1-11 | tt7 A1-3 |
Arabidopsis tt5 mutants (tt5-1 allele) transformed with the maize CHI1 gene (Grotewold and Peterson, 1994) under control of the CaMV 35S promoter also showed full restoration of both cotyledon pigmentation in the low-nitrogen media assay and seed coat pigmentation (Table I and Fig. 2A,tt5). The 35S::CHI1 construct also induced the accumulation of UV-absorbing compounds that block the fluorescence oftt5 seeds (not shown).
Similar experiments conducted using the maize A1 gene, encoding DFR (Schwarz-Sommer et al., 1987), under the control of the CaMV 35S promoter demonstrated that this construct was capable of restoring the pigmentation of tt3 mutant plants and seeds (Table I and Fig. 2A). Unlike tt4 and tt5 seeds,tt3 seeds accumulate UV-absorbing compounds and are not fluorescent. Thus, the 35S::A1 construct did not alter the fluorescence of these mutant seeds (not shown).
Expression of all of the transgenes was investigated in wild type and corresponding transparent testa mutant seedlings grown in low-nitrogen media, and, where appropriate, mature plants. Most of the transgenic lines investigated (Table I) show accumulation of C2, CHI1, or A1 transcripts, although at variable levels (Fig. 2B). Landsbergerecta C2-3 seedlings show very low levels of C2 expression, evident only with very long exposures (not shown). In a similar manner, CHI1 transcripts in tt5 CHI-2 seedlings could not be detected even with long exposures (Fig. 2B), despite the complemented phenotype of tt5 CHI-2 seedlings (Fig. 2B), suggesting that low levels of CHI1 expression are sufficient for full phenotypic complementation of the tt5 mutation. Together, these results demonstrate that the maize C2, CHI1, andA1 genes encode flavonoid biosynthetic enzymes that are fully functional in Arabidopsis and further provide direct evidence that the Arabidopsis transparent testa mutant phenotypes result from the absence of these enzymatic activities.
Comparison of the Flavonoids Accumulated in Wild-Type, Mutant, and Transgenic Plants
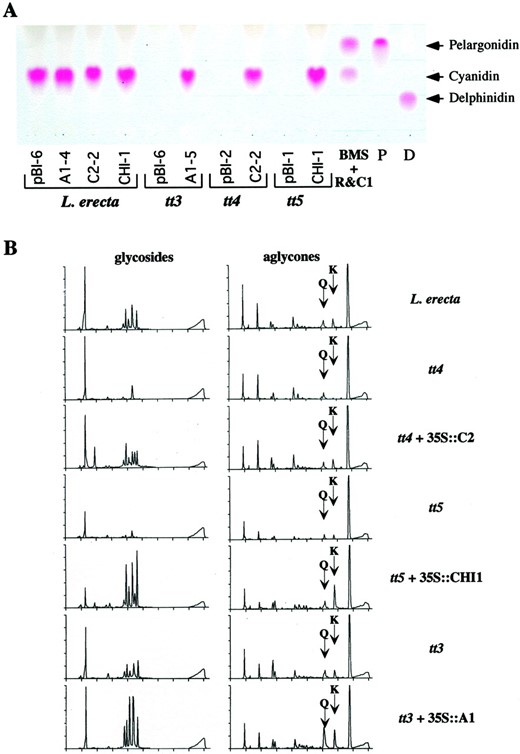
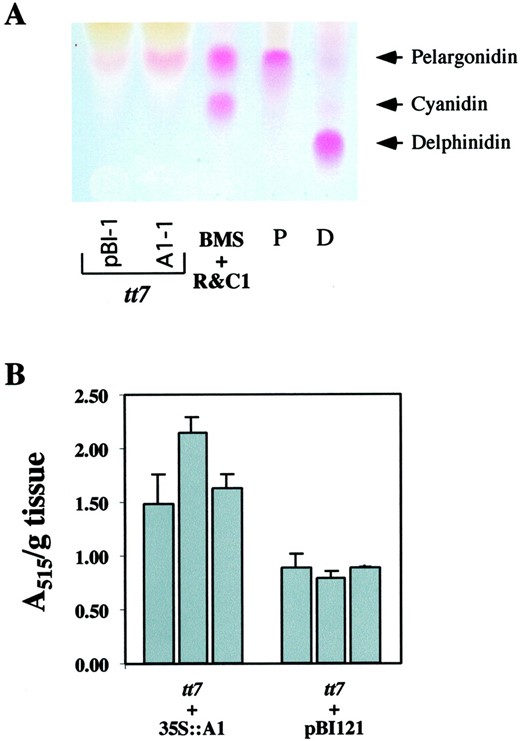
Anthocyanidin and flavonoid accumulation in wild type and transgenic Arabidopsis plants. A, Thin-layer chromatography (TLC) analysis of the anthocyanidins that accumulate in wild-type (Landsberg erecta) or mutant tt3, tt4, or tt5 Arabidopsis seedlings grown on low-nitrogen media expressing the maize C2, CHI, or A1genes. BMS R&C1 corresponds to the anthocyanidins found in BMS cells expressing the R and C1 regulators of anthocyanin biosynthesis (Grotewold et al., 1998). The pelargonidin standard (P) was obtained from geranium (Pelargonium cv Salmon Mbl Mix) flowers, and the delphinidin (D) from lisianthus (Eustoma grandiflorum Grise variety Royal Violet) flowers (see “Materials and Methods”). B, HPLC analysis of flavonoids present in methanol extracts of equivalent amounts (wet weight) of leaves from 4-week-old wild-type (Landsberg erecta) or mutant Arabidopsis, in the presence or absence of the maize C2, CHI1, orA1 genes. The left column shows the accumulation of the non-hydrolized glycosides, and the right column shows the accumulation of hydrolized aglycones. The mobility of Q or K standards is indicated by arrows.
To further characterize the flavonoids synthesized by the Arabidopsis mutants expressing the maize genes, HPLC experiments were conducted using both acid hydrolyzed and non-hydrolyzed ethanol extracts from leaves of 4-week-old plants (Fig. 3B). Expression of orthologous maize genes in tt4 and tt5 mutants restores the accumulation of K and Q (Fig. 1; compare tt4with tt4 + 35S::C2 and tt5 withtt5 + 35S::CHI in Fig. 3B). As previously reported (Pelletier et al., 1999), both wild-type and tt3 plants accumulate modest amounts of K and Q, but these are absent intt4 and tt5 mutants (Fig. 3B). The peaks with mobility similar to Q in the tt4 mutant, or with Q and K in the tt5 mutant migrate slightly different than the K and Q standards, and their absorption spectra is different from those of K and Q (not shown). However, there are a few remarkable differences in the quality or relative quantity of the flavonoids present in transgenic plants when compared with wild-type plants. First, the expression of C2 in tt4 plants resulted in the appearance of six, rather than five, glycoside peaks in the non-hydrolyzed extracts (compare tt4 + 35S::C2 and Landsbergerecta, Fig. 3B). Second, expression of CHI1 intt5 plants resulted in a higher K/Q ratio than that found in wild-type plants (compare tt5 + 35S::CHI1 with Landsberg erecta, Fig. 3B). Finally, the expression of A1 in the tt3 mutant resulted in an increased accumulation of both K and Q when compared with either the wild-type or to tt3mutant plants (compare tt3 + 35S::A1 withtt3 or Landsberg erecta, Fig. 3B). Whether these unexpected findings are a consequence of slight differences in the developmental stage of the wild-type and mutant plants, which is known to affect the accumulation of both Q and K (Pelletier et al., 1999), or unique properties of the maize enzymes remains unclear.
Overexpression of C2, CHI1, andA1 Does Not Significantly Increase Anthocyanidin Accumulation
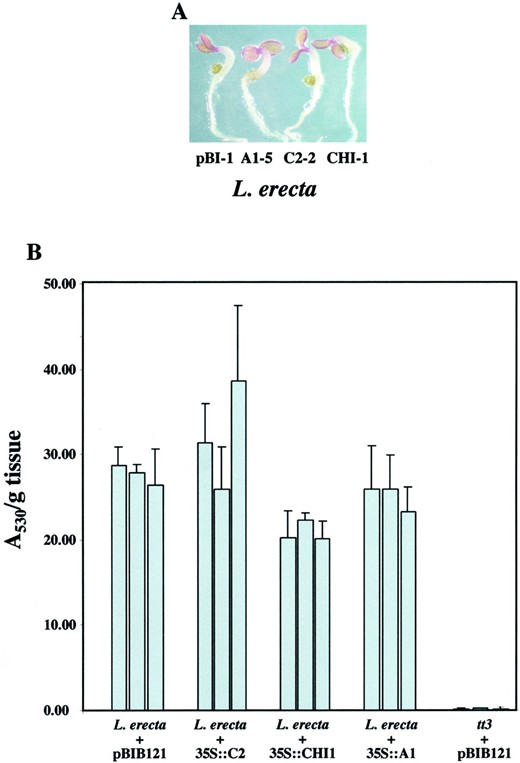
Expression of the maize flavonoid biosynthetic genes in wild-type Arabidopsis seedlings does not result in an increase of anthocyanidin accumulation. A, Wild-type (Landsbergerecta) Arabidopsis seedlings expressing the maizeA1 (A1-5), C2 (C2-2), or CHI1 (CHI-1) genes show similar levels of pigment accumulation as compared with wild-type seedlings carrying the empty binary vector (pBI-1) when grown in low-nitrogen media. B, Comparison of cyanidin accumulation in seedlings of three lines of wild-type (Landsberg erecta) Arabidopsis expressing the maize genes. The error bars correspond to the sd of triplicate measurements for each line.
The accumulation of the anthocyanidin pigments was quantified using spectrophotometry. The A 530 oftt3 seedlings grown in low-nitrogen Murashige and Skoog media is close to zero (Fig. 4B), confirming that this wavelength is measuring primarily anthocyanidin pigments. NormalizedA 530 values (Fig. 4B) indicate that the ectopic expression of the maize C2, CHI1, orA1 genes in wild-type Arabidopsis did not result in a statistically significant increase in anthocyanidin accumulation (compare Landsberg erecta + pBIB121 with Landsbergerecta + 35S::C2, Landsberg erecta + 35S::CHI1, and Landsberg erecta + 35S::A1 in Fig. 3B). There was also little variation in anthocyanidin accumulation among plants transformed with the same gene but derived from independent integration events (compare among lines in Landsberg erecta + 35S::C2, Landsbergerecta + 35S::CHI1, and Landsbergerecta + 35S::A1 in Fig. 3B).
It is surprising that 35S::CHI1 expressing lines had a significant decrease in anthocyanidin accumulation (P = 0.0001), resulting in A 530 values approximately 75% of those in the wild type. This unexpected decrease in cyanidin accumulation might be due to a partial inhibitory effect of the maize CHI1 enzyme on the endogenous CHI; for example, by competing for interaction with CHS (Burbulis and Winkel-Shirley, 1999). The observed formation of an increased amount of K relative to Q in plants expressing the maize CHI1 enzyme (Fig. 3B) alternatively may limit the available DHQ necessary for cyanidin formation (Fig. 1). However, it is also possible that wild-type plants expressing maize CHI1 grow at a slightly different rate than wild-type plants, which could alter the accumulation of anthocyanidins relative to tissue mass.
Accumulation of Pelargonidin Pigments in Flavonoid 3′ Hydroxylase (F3′H) Mutants
Wild-type Arabidopsis seedlings accumulated mainly cyanidin pigments when grown on low-nitrogen media (Fig. 3A). The absence of detectable pelargonidin under these conditions suggests that the Arabidopsis DFR enzyme has a high degree of preference for DHQ over DHK (Fig. 1). The fact that mutations in the F3′H enzyme responsible for the conversion of DHK to DHQ results in a pale-brown transparent testa phenotype (F3′H is encoded by the TT7 locus [Koorneef et al., 1982; Schoenbohm et al., 2000]) suggests that pigment formation does not proceed very efficiently using DHK. In contrast, maize cells expressing the A1-encoded DFR enzyme accumulate both pelargonidin and cyanidin (Fig. 3A), indicating that both compounds can be efficiently utilized by A1. However, expression of A1 in wild-type or tt3 Arabidopsis seedlings did not result in pelargonidin accumulation (Fig. 3A), despite the clear ability of A1 to utilize DHK as a substrate in maize.
Accumulation of pelargonidin in F3′H mutant seedlings. A, TLC analysis of anthocyanidins extracted fromtt7 mutant seedlings expressing the maize A1 gene (tt7 A1-1) or an empty vector (tt7 pBI-1). The conditions and standards are identical to Figure 3. B, Comparison of pelargonidin accumulation in seedlings of three tt7 lines (Table I) expressing the maize A1 gene (tt7 + 35S::A1) or the empty vector (tt7 + pBI121). The error bars correspond to the sd of triplicates for each line.
To quantify this difference, the amount of pelargonidin that accumulated in these seedlings was estimated using the characteristic absorbance peak of pelargonidin at 515 nm (Fig. 5B). Threett7 lines expressing the maize A1 gene (Fig. 2D and results not shown) were analyzed, each one in triplicate, and compared with tt7 lines carrying the empty pBIB121 vector. As evident in Figure 5B, the tt7 lines expressing A1 accumulate approximately twice as much pelargonidin than tt7mutants. This level of pelargonidin accumulation is not sufficient for either visual pigmentation in our seedling assay or to provide visible pigmentation in the seed coat (data not shown), although the increased accumulation of pelargonidin based uponA 515 measurements was highly significant (P = 3 × 10−8). The variation observed in the level of anthocyanidin accumulation between different 35S::A1 tt7 lines (Fig. 5B) is not significant based upon a two-factor ANOVA (P = 0.055).
These findings are consistent with the hypothesis that the maize DFR enzyme encoded by A1 has a higher preference for DHK (Fig.1) relative to the Arabidopsis enzyme, and that the absence of pelargonidin in wild-type plants overexpressing A1 is a consequence of a competition for DHK between A1 and TT7, flavonol synthase, or both (Fig. 1). However, we cannot exclude the possibility that the elevated accumulation of pelargonidin in A1-expressing tt7 mutant plants reflects the overexpression of A1, which would suggest that DFR activity is limiting for pelargonidin accumulation under the conditions tested, but not for cyanidin formation, as discussed above. Together, these findings indicate that both the maize and Arabidopsis DFR enzymes can utilize DHK as a substrate, but fail to do so in Arabidopsis plants when a functional F3′H is present.
DISCUSSION
The origin of the anthocyanin biosynthetic pathway precedes the divergence of monocots and dicots (Stafford, 1991), providing an ideal opportunity to investigate the functional conservation of enzymes involved in the formation of specific secondary metabolites. We have demonstrated that the products of the maize C2,CHI1, and A1 genes can fully complement Arabidopsis tt4, tt5, and tt3 mutants, providing evidence that enzymes involved in secondary metabolite accumulation are exchangeable between distantly related plants.
The function of the Arabidopsis TT4, TT5, andTT3 genes, encoding CHS, CHI, and DFR enzymes, has been inferred from the molecular analysis of multiple alleles (when available), from the effects on the expression of the corresponding mRNAs and proteins, and from the accumulation of particular intermediates in the pathway (Shirley et al., 1992, 1995; Burbulis et al., 1996). To our knowledge, none of these mutations has been complemented with the corresponding wild-type genes from Arabidopsis. The finding that the maize C2, CHI1, and A1 enzymes complement thett4-1, tt5-1, and tt3-1 alleles unequivocally confirms the previous assignment of tt4 to CHS, tt5 to CHI, and tt3 to DFR.
Previous studies demonstrated that the maize DFR enzyme encoded byA1 is functional in dicots (Meyer et al., 1987). The expression of the maize A1 gene in petunia plants mutant for the F3′H and F3′5′H genes represented the first application of transgene technology to modify flower pigmentation. Petunia flowers expressing A1 accumulated brick-red/orange pelargonidin pigments absent from wild-type flowers, which accumulate the cyanidin or delphinidin pigments. These findings indicated that the maize DFR enzyme encoded byA1 has the potential to use both DHK and DHQ as substrates (Fig. 1). BMS cells expressing the R&C1 regulators of anthocyanin biosynthesis (Grotewold et al., 1998) also induce the accumulation of both pelargonidin as well as cyanidin-derived pigments (Fig. 3A).
Wild-type Arabidopsis seedlings accumulate only cyanidin (Fig. 3A). It is interesting that cyanidin is also the only anthocyanidin found when the maize A1 gene is expressed in Arabidopsis, whether or not the endogenous DFR enzyme is present (Fig. 3A). Northern-blot experiments indicate that A1 is expressed in wild-type andtt3 plants at similar levels (Fig. 2D). However, in the absence of the F3′H activity encoded by TT7, pelargonidin anthocyanidins accumulate, indicating that Arabidopsis DFR has the capability to utilize DHK as a substrate. The expression of A1 in thett7 mutants results in a further increase in the accumulation of pelargonidin-derived pigments, consistent with the findings in maize plants (Styles and Ceska, 1975), maize BMS cells (Fig. 3A), and petunia flowers (Forkmann and Ruhnau, 1987) that this enzyme can utilize DHK as a substrate. The absence of pelargonidin pigments in either wild-type or A1-expressing Arabidopsis plants indicates that the conversion of DHK to DHQ by F3′H is much more efficient than the utilization of DHK as a substrate by either DFR enzyme. The substrate specificity of the Gerbera DFR was altered recently by a single amino acid change (Johnson et al., 2001), suggesting that very related DFR enzymes can have a strong preference of one substrate over another. An alternative explanation for our findings is that the TT7-encoded F3′H enzyme plays a key role in substrate channeling in the complex formed by the Arabidopsis flavonoid biosynthetic enzymes (Burbulis and Winkel-Shirley, 1999;Winkel-Shirley, 1999). If so, it appears that the maize A1 protein can participate in the metabolic channel in a manner similar to the DFR encoded by the endogenous Arabidopsis TT3 gene.
The maize C2 and A1 gene have been cloned with the aid of mutant alleles that abolish anthocyanin accumulation (Wienand et al., 1986; Schwarz-Sommer et al., 1987), whereas the maizeCHI1 gene was cloned by homology to CHI enzymes from other plants (Grotewold and Peterson, 1994). No mutations in any of the two or three CHI genes present in various maize inbred lines have so far been described. Moreover, in striking contrast to the Arabidopsis tt5 mutant, maize BMS cells lackingCHI1 mRNA as well as CHI enzymatic activity accumulate high levels of flavonoids when the other enzymes in the flavonoid biosynthetic pathway are induced by expression of the P or R&C1 regulators (Grotewold et al., 1998). These findings suggest thatCHI1 might not encode a functional CHI, and raise further questions about the necessity of a CHI activity for flux through the flavonoid pathway. In addition, these results conclusively demonstrate that the absence of flavonoids that characterize tt5 are solely due to a mutation in the gene encoding CHI. It is unclear why CHI is necessary for flavonoid accumulation in Arabidopsis but not in maize, given the spontaneous isomerization of chalcones to flavanones in maize cells and in vitro (Grotewold et al., 1998).
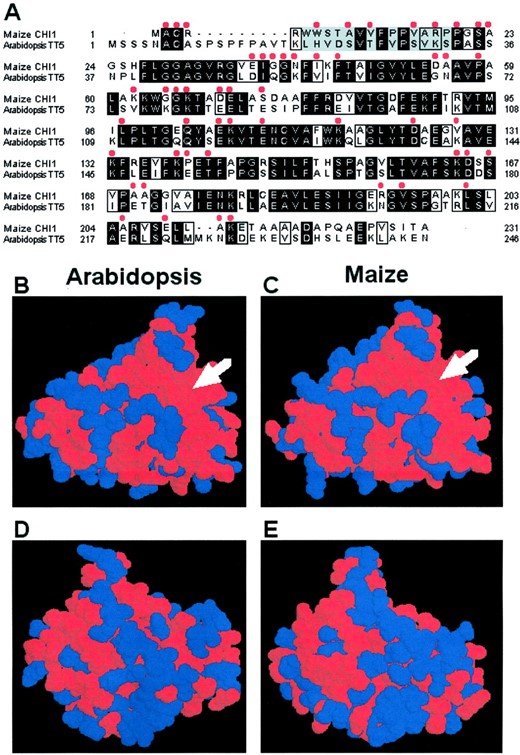
Recent findings indicate that Arabidopsis flavonoid biosynthetic enzymes form large macromolecular complexes through specific protein-protein interactions (Burbulis and Winkel-Shirley, 1999), suggesting that the flux of intermediates into different products could be controlled by the formation of distinct complexes (Winkel-Shirley, 1999). The potential for formation of similar complexes in other plants has not been rigorously examined, although in vitro experiments have failed to identify an alfalfa (Medicago sativa) CHS-CHI complex (Jez et al., 2000). The observation that all three maize flavonoid biosynthetic enzymes tested here can function efficiently in Arabidopsis argues that they can either participate in the formation of the observed macromolecular complexes or that the formation of these enzymatic complexes has little influence in the formation of anthocyanins. The high degree of sequence conservation (89% identity) between the maize and Arabidopsis CHS makes it feasible that the maize enzyme physically interacts with the Arabidopsis CHI and DFR enzymes, like the Arabidopsis CHS (Burbulis and Winkel-Shirley, 1999). However, the limited conservation between the maize and Arabidopsis CHI (58% identity) and DFR (59% identity) proteins opens the question as to whether residues involved in protein-protein interaction are conserved in these proteins.
The existence of a second Arabidopsis gene on chromosome 5 encoding a protein homologous to CHI (designated AT5g66220 or K2A18.30) raises additional questions. The AT5g66220 protein has a greater identity to the TT5-encoded CHI (68% identity) than either Arabidopsis protein exhibits to the maize CHI1 protein (E.L. Braun and E. Grotewold, unpublished data). The AT5g66220 protein clearly does not provide an activity that can complement the phenotypes of thett5 mutants. It is not clear whether the phenotype oftt5 mutants reflects insufficient expression of AT5g66220 or the absence of a specific biological activity for the AT5g66220 protein. However, the absence of complementation from this protein illustrates the difficulty associated with predicting the functionality of proteins involved in secondary metabolism using homology searches. Likewise, the ability of the maize CHI1 protein to complement thett5 mutation demonstrates the power of genetic approaches to elucidate the actual biochemical activity of these proteins, even when they are isolated from evolutionarily divergent plant species.
Comparison of structural models for the maize and Arabidopsis CHI enzymes. A, Alignment of the maize CHI1 with the Arabidopsis TT5-encoded CHI proteins. Identical residues are highlighted with a black background and conservative substitutions are boxed. The β1a, β1b, and β2β-strands suggested as providing a potential protein-protein interaction surface (Jez et al., 2000) are highlighted in gray. Residues that are clearly exposed to the solvent (defined here as having either more than 10 Å2exposed or being more than 33% solvent accessible as the same residue in an unfolded peptide) are indicated with red dots over the amino acid. B, Space filling model of the Arabidopsis CHI protein with the substrate-binding site indicated with an arrow. Residues that are identical in the maize and Arabidopsis proteins are indicated in red. C, Space filling model of the maize CHI1 protein, view is identical to B. D, Space filling model of the Arabidopsis CHI protein showing the opposite face. E, Space filling model of the maize CHI1, view is identical to panel D. The N- and C-terminal regions of the proteins were not included in the models.
It was proposed that the Arabidopsis CHI enzyme is posttranslationally modified through a thioester linkage involving one or more of the three Cys residues present, and that this modification might be involved in the association of the multiprotein complex with the endoplasmic reticulum (Burbulis and Winkel-Shirley, 1999). If so, this would suggest that a similar modification might occur to the maize CHI1 protein. The maize CHI1 enzyme has two of these three Cys residues conserved (Fig. 6A), with only the Cys at position 3 in the maize protein predicted to be solvent exposed, and therefore available for a modification.
Together, the findings presented here provide strong evidence that at least some enzymes involved in secondary metabolic pathways conserved between monocots and dicots are exchangeable between these two divergent groups of flowering plants. The qualitative and quantitative differences in the accumulation of particular flavonoids when the maize enzymes are expressed in Arabidopsis suggest that modest differences in their substrate preferences may exist, providing both powerful tools and remarkable challenges for the engineering of novel metabolites in specific plants. The recent demonstration that a soybean (Phaseolus vulgaris) isoflavone synthase, by itself or in the presence of a soybean chalcone reductase, is capable of inducing the accumulation of soybean isoflavonoids (genistein and daidzein) in maize, tobacco (Yu et al., 2000), and Arabidopsis (Jung et al., 2000), provides further evidence of the efficacy of the heterologous expression of biosynthetic enzymes for metabolic engineering. The findings presented here also indicate the value of the Arabidopsis flavonoid biosynthetic mutants as a convenient system to assay the activity of biosynthetic genes from other plants. In fact, the complementation of Arabidopsis transparent testa mutants should be a useful system for establishing the function of uncharacterized open reading frames with homology to flavonoid biosynthetic enzymes when those genes actually encode proteins with a role in flavonoid biosynthesis.
MATERIALS AND METHODS
Plant/DNA Materials and Plant Growth Conditions
All the transparent testa (tt) mutants were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus). All the plants used in these studies are in the Landsberg erecta genetic background. Seeds were germinated on one-half-strength Murashige and Skoog (Murashige and Skoog, 1962) agar plates (Murashige and Skoog salts [GIBCO/BRL, Grand Island, NY], pH adjusted to 5.7 with potassium hydroxide, 0.5% [w/v] agar) containing 1% (w/v) Suc. Murashige and Skoog basal medium without a nitrogen source was from GIBCO/BRL (formula no. 97-5068EC). For antibiotic selection, seeds were grown in one-half-strength Murashige and Skoog plates with 12 μg mL−1 of hygromycin. After 1-week selection in plates, hygromycin-resistant plants were transplanted to flats containing M360 soil (Hummert Seed Co., Earth City, MO), covered with plastic domes and kept in the cold room at 4°C for 2 d to break seed dormancy and to synchronize the germination of the seeds. After 2 d, the flats were transferred to controlled growth chambers operating at 22°C and 50% to 80% relative humidity under continuous light. One week after planting, the transparent covers were removed and the seedlings were watered as needed.
The full-length maize (Zea mays) C2 (Wienand et al., 1986), CHI1 (Grotewold and Peterson, 1994), and A1 (Schwarz-Sommer et al., 1987) cDNAs were cloned into the SmaI site of the pBIB121 binary vector, so that the corresponding genes were placed downstream of the CaMV 35S promoter. The resulting plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. A. tumefaciens was grown to stationary phase, harvested by centrifugation, and resuspended in 2 volumes of water with 0.2% (v/v) Silwet l-77 (Lehle Seeds, Round Rock, TX). Transformation of Arabidopsis plants was done by the vacuum infiltration method (Bechtold et al., 1993), with transformation efficiencies of 1% to 2% of the harvested seeds. Transformed T1 seedlings were selected on medium containing 30 μg mL−1 hygromycin and 500 μg mL−1carbenicillin. Seedlings with two leaves were then transferred to soil and grown at 22°C in a growth room with a 16-h-light/-dark cycle.
Extraction and Analysis of Anthocyanidins
T2 seeds of three independent transgenic lines were germinated in triplicate on low-nitrogen medium. After 10 d in high light, seedlings were harvested, pigments were extracted in 1.5 mL of 1% (v/v) HCL in methanol, and 1.0 mL of distilled water was added. Chlorophyll was separated from the anthocyanidins by back extraction with chloroform. Anthocyanidins were quantified using theA 530 for cyanidin, or at 515 nm (A 515) for pelargonidin. This absorbance value was normalized for the fresh weight of tissue used. Differences between and among groups of transformants were assessed using the two-factor analysis of variance (ANOVA) either with replication (for differences between plants transformed with different genes) or without replication (for differences within sets of plants transformed with the same gene) as implemented in Microsoft Excel 98 (Microsoft Corporation, Redmond, WA). Extracts from flowers of lisianthus (Eustoma grandiflorum Grise variety Royal Violet) and geranium (Pelargonium cv Salmon Mbl Mix) were used as standards for delphinidin and pelargonidin, respectively.
HPLC Analysis of Flavonoids and TLC Analysis of Anthocyanidins
A sample of 25 mg of tissue (fresh weight) was ground to a fine suspension in 0.2 mL of 80% (v/v) ethanol in a polypropylene microcentrifuge tube (1.5 mL) with a polypropylene pestle (Kontes Glass Co., Vineland, NJ) at 600 rpm. The extracts were centrifuged (13,000g for 3 min) to pellet insoluble debris and the supernatants used for HPLC analyses. Acid hydrolysis of ethanol extracts to produce the aglycones was carried out as previously described for the hydrolysis of methanol extracts (Burbulis et al., 1996). Injections of 20 μL were made onto a 4.6- × 250-mm Hibar Ec Cartridge containing Merck Lichrosorb RP-18 10 MM C18 reverse phase packing (Alltech Associates, Deerfield, IL), with a guard column containing the same matrix used to protect the analytical column. Chromatography was performed at 25°C with a low-pressure pump (Spectroflow model 430, Applied Biosystems, Foster City, CA) to form a linear gradient of 0% to 55% (v/v) acetonitrile at pH 3.0 in water for 25 min, followed by a step increase to 100% (v/v) acetonitrile, which was held for 2 min before a step return to HPLC grade water (adjusted to pH 3.0 with phosphoric acid). A flow rate of 1.5 mL min−1 was used. The overall separation time, including the 3-min water wash, was 25 min. The spectrometric detection was carried out at 255 nm. The Q and K standards were obtained from Fluka BioChemika (Buchs, Switzerland).
For TLC analyses, anthocyanidins were extracted by crushing the tissue with a plastic pestle in 2 m HCl, and boiling the material for 20 min. After extraction, four to five drops (approximately 200 μL) of isoamylic alcohol were added, and after mixing, the organic phase was separated from the aqueous. Cellulose TLC plates (Merck, Dorset, UK) were run in a mobile phase formed by water-formic acid-HCL (10-30-3).
RNA Extraction and Northern-Blot Analyses
Total RNA was isolated utilizing the Trizol reagent (Life Technologies Inc., Gaithersburg, MD), according to the manufacturer's instructions, either from 10-d-old seedlings or from mature plants. One to 10 μg of total RNA was separated on formaldehyde-containing 1% (w/v) agarose gels (Sambrook et al., 1989) and probed with different agarose-purified DNA fragments.
Molecular Modeling and Sequence Alignments
Sequence alignments were performed using ClustalW (Thompson et al., 1994) and molecular models were generated using WHAT IF (Vriend, 1990) with the 1.85-Å structure of CHI from alfalfa (Medicago sativa; Jez et al., 2000). To assess the conservation of buried and exposed residues, aligned residues were assigned to either the buried or exposed category as described by Goldman et al. (1998). The null hypothesis that categories in a 2 × 2 contingency table (generated by further categorizing these residues as identical or different in comparisons between the maize and Arabidopsis proteins) are homogeneous was evaluated using the χ2 test.
ACKNOWLEDGMENTS
We thank Ronald Koes for teaching us how to separate anthocyanins by TLC, Brenda Shirley for helpful discussions on the manuscript and for sharing unpublished information, Iris Meier for comments on the manuscript, Terry Graham for allowing us to use his HPLC equipment, and Gloria Coruzzi for providing the Murashige and Skoog media without nitrogen. We also thank two anonymous reviewers for their comments that significantly improved the manuscript. We thank the Arabidopsis Biological Resource Center for the mutant Arabidopsis stocks. We are very grateful to J. Marcela Hernandez for excellent technical assistance.
LITERATURE CITED
Author notes
This work was supported in part by the National Science Foundation (grant nos. MCB–9974474 and MCB–9896111 to E.G.) and by the U.S. Department of Agriculture (postdoctoral fellowship no. USDA 1999–01582 to E.L.B.).
Corresponding author; e-mail grotewold.1@osu.edu; fax 614–292–5379.