-
PDF
- Split View
-
Views
-
Cite
Cite
Sandra L MacKenzie, Matt C Wyatt, Robert Schuff, Jessica D Tenenbaum, Nick Anderson, Practices and perspectives on building integrated data repositories: results from a 2010 CTSA survey, Journal of the American Medical Informatics Association, Volume 19, Issue e1, June 2012, Pages e119–e124, https://doi.org/10.1136/amiajnl-2011-000508
Close - Share Icon Share
Abstract
Clinical integrated data repositories (IDRs) are poised to become a foundational element of biomedical and translational research by providing the coordinated data sources necessary to conduct retrospective analytic research and to identify and recruit prospective research subjects. The Clinical and Translational Science Award (CTSA) consortium's Informatics IDR Group conducted a survey of 2010 consortium members to evaluate recent trends in IDR implementation and use to support research between 2008 and 2010. A web-based survey based in part on a prior 2008 survey was developed and deployed to 46 national CTSA centers. A total of 35 separate organizations completed the survey (74%), representing 28 CTSAs and the National Institutes of Health Clinical Center. Survey results suggest that individual organizations are progressing in their approaches to the development, management, and use of IDRs as a means to support a broad array of research. We describe the major trends and emerging practices below.
Background
Growing adoption of electronic health records (EHRs) in recent years has given rise to an increased opportunity for ‘secondary use,’ that is, leveraging the data collected through the course of clinical care for purposes of research and quality improvement.1 In order to enable these tasks, a number of academic health centers have developed enterprise data warehouses based on, but distinct from, the transaction-based clinical care system. Described alternatively as clinical data warehouses, integrated data warehouses or integrated data repositories (IDRs, the term we will use in this document), development of these systems at the institutional level can be a considerable challenge. IDRs combine disparate data types from a range of clinical applications, including home-grown, commercial, and customized systems. They can extend across multiple information boundaries, from department to institution to governmental reporting, as well as between disease domains. And they are intended to support a range of heterogeneous end-users and functions, including research, clinical operations, and quality assurance/quality improvement.
There has been an emphasis in recent years on translational research, that is, translating laboratory discoveries into clinical guidelines and ultimately into clinical practice. Also part of translational research, although less frequently addressed, is translating in the other direction using clinical practice to inform research.2 In 2006, the NIH (National Institutes of Health) created the Clinical and Translational Science Award (CTSA) funding mechanism to advance human health by transforming the research and training environment at a national level.3 Sixty funded CTSA sites form the CTSA consortium, providing infrastructure not only to transform biomedical research within the respective institutions, but also to enable the sharing of best practices and lessons learned, within the consortium and beyond. The CTSA consortium now includes many leading academic medical centers and large healthcare delivery organizations in the USA. The consortium infrastructure provided an opportunity to survey IDR-related efforts in a systematic way, thus enabling a national perspective on implementation efforts for advancing secondary use of clinical data for research.
With the initial goals of fostering collaborative work and sharing best practices, the IDR Working Group sought first to systematically define and evaluate what an ‘integrated data repository’ represented to different sites and the functional role that IDRs serve. Later inquiries sought to identify what common challenges were being encountered and how these challenges evolved over time, as well as the lessons learned. In 2007, shortly after the first cohort of 12 CTSAs had been awarded, an initial survey was created to solicit feedback on the evolving definitions of an IDR and to identify CTSAs with clinical data repositories that were being accessed for research data. The follow-up 2008 survey was designed to provide an updated view of the number and scope of IDRs. It also served to establish a baseline for measuring changes in key IDR characteristics. The 2010 IDR survey (the focus of this paper) was designed to achieve the following goals:
Gain insight into the adoption and use of IDRs by CTSA consortium members
Identify trends since the 2008 survey
Identify opportunities for sharing tools, knowledge and resources
Identify areas for future exploration, such as readiness for larger scale data sharing between CTSAs.
The survey results and perspectives in this paper should be of broad interest to institutions considering the deployment of research IDRs in an academic health system environment, or to those pursuing expanded use of existing IDRs.
Methods
Survey development
The surveys were designed both to collect information and to guide future inquiry. Many of the questions from the 2008 survey were based on the Health Data Warehousing Association Tools Inventory.4 Using the 2008 survey as a base, we (RS, MW, SM) incorporated additional questions to address topics such as repository size, annual growth, number of unique patients, system architecture, front-facing business intelligence tools, and planned enhancements. The end result was a 77-item survey for 2010 (see online supplementary appendix A). Survey questions were organized in sections as shown in table 1.
Survey sections and descriptions
| Section . | Description . |
|---|---|
| General IDR status | Characteristics of the IDR, such as size, annual growth, number of unique patients, if a single or multi-institution IDR |
| Data sources supporting the IDR | Types of data incorporated in an organization's IDR, for example, diagnoses, visit details, lab results, meds, patient charges |
| Technical architecture and staffing | Software tools supporting the IDR infrastructure including RDBMS, extract transformation load applications, front-end reporting/BI tools, and level of IDR support staffing |
| Data access and compliance | Methods employed for access to IDR data based on whether info desired is aggregate data, de-identified, or fully identifiable patient data |
| Clinical systems | Transaction based clinical systems in an organization, whether currently supplying data to the IDR or not, for example, inpatient and/or ambulatory EMR, lab systems, ER systems, more |
| Project experiences | Obstacles encountered during IDR development, lessons learned, continuing challenges, and planned enhancements |
| Section . | Description . |
|---|---|
| General IDR status | Characteristics of the IDR, such as size, annual growth, number of unique patients, if a single or multi-institution IDR |
| Data sources supporting the IDR | Types of data incorporated in an organization's IDR, for example, diagnoses, visit details, lab results, meds, patient charges |
| Technical architecture and staffing | Software tools supporting the IDR infrastructure including RDBMS, extract transformation load applications, front-end reporting/BI tools, and level of IDR support staffing |
| Data access and compliance | Methods employed for access to IDR data based on whether info desired is aggregate data, de-identified, or fully identifiable patient data |
| Clinical systems | Transaction based clinical systems in an organization, whether currently supplying data to the IDR or not, for example, inpatient and/or ambulatory EMR, lab systems, ER systems, more |
| Project experiences | Obstacles encountered during IDR development, lessons learned, continuing challenges, and planned enhancements |
BI, business intelligence; EMR, electronic medical record; ER, emergency room; IDR, integrated data repository; RDBMS, relational database management system.
Survey sections and descriptions
| Section . | Description . |
|---|---|
| General IDR status | Characteristics of the IDR, such as size, annual growth, number of unique patients, if a single or multi-institution IDR |
| Data sources supporting the IDR | Types of data incorporated in an organization's IDR, for example, diagnoses, visit details, lab results, meds, patient charges |
| Technical architecture and staffing | Software tools supporting the IDR infrastructure including RDBMS, extract transformation load applications, front-end reporting/BI tools, and level of IDR support staffing |
| Data access and compliance | Methods employed for access to IDR data based on whether info desired is aggregate data, de-identified, or fully identifiable patient data |
| Clinical systems | Transaction based clinical systems in an organization, whether currently supplying data to the IDR or not, for example, inpatient and/or ambulatory EMR, lab systems, ER systems, more |
| Project experiences | Obstacles encountered during IDR development, lessons learned, continuing challenges, and planned enhancements |
| Section . | Description . |
|---|---|
| General IDR status | Characteristics of the IDR, such as size, annual growth, number of unique patients, if a single or multi-institution IDR |
| Data sources supporting the IDR | Types of data incorporated in an organization's IDR, for example, diagnoses, visit details, lab results, meds, patient charges |
| Technical architecture and staffing | Software tools supporting the IDR infrastructure including RDBMS, extract transformation load applications, front-end reporting/BI tools, and level of IDR support staffing |
| Data access and compliance | Methods employed for access to IDR data based on whether info desired is aggregate data, de-identified, or fully identifiable patient data |
| Clinical systems | Transaction based clinical systems in an organization, whether currently supplying data to the IDR or not, for example, inpatient and/or ambulatory EMR, lab systems, ER systems, more |
| Project experiences | Obstacles encountered during IDR development, lessons learned, continuing challenges, and planned enhancements |
BI, business intelligence; EMR, electronic medical record; ER, emergency room; IDR, integrated data repository; RDBMS, relational database management system.
The 2010 survey was created using REDCap (Research Electronic Data Capture), a widely used web-based platform for survey deployment.5 The majority of the survey questions asked for structured responses. A subset of questions provided the option for free text answers in order to capture qualitative or unanticipated data. Use of free text was deemed appropriate given that the intent of the survey was to enable collection of novel and unanticipated experiences, rather than to be a rigorously validated scientific instrument.
Survey deployment and analysis
A message was sent to the CTSA Informatics Directors email distribution list describing the survey and requesting contact information for the appropriate point person(s) for IDRs associated with their core CTSA institution and respective partner organizations. The Informatics Directors list represents the voting membership of the Informatics Key Function Committee (IKFC), each of whom respectively represents the informatics component of his or her particular CTSA site, with one voting member per site. The individuals identified were contacted and emailed survey URLs as their information was received. Fourteen of the directors completed the surveys themselves; the remainder were completed by delegates. A follow-up message was sent 2 weeks later to those who had not responded. The survey was administered from May to July 2010 to 47 organizations representing 36 CTSAs (eg, Memorial Sloan-Kettering Cancer Center and Weill Cornell Medical College are two separate organizations within a single CTSA) as well as the NIH Clinical Center. Survey completion was tracked and follow-up notifications were provided to those who had not completed the survey. Of the 47 organizations receiving a survey invitation, 35 separate organizations completed a survey (74%), representing 28 CTSAs and the NIH Clinical Center. Seventeen of the 35 (49%) were institutions who had responded in 2008 (56% of the 30 who respond in 2008). Twelve of those 17 replies were by the same respondents who had completed the 2008 survey; the remainder were different individuals. All survey responses received were included in our analysis. During analysis, free text responses took one of three forms: numerical (eg, population counts, data size, date ranges), lists of items (eg, products/vendors, terminology variants), and narrative text responses. Numerical/date responses were normalized to common units and ranges. Lists of text items were parsed and clustered by theme. All responses to free text questions were clustered according to theme by SM, with independent verification by MW and RS. If a response to a free text question covered multiple categories, each was counted as a separate item (eg, ‘staffing and funding levels' was counted as staffing levels and as funding levels). Results were presented to the CTSA IDR Group in December 2010 via an online webinar.
Results
Survey results can be found in online supplementary appendix B. The responses have been aggregated to prevent identification of individual institutions. Overall, the 2010 CTSA IDR survey shows a growing prevalence of IDRs and provides insight into the evolving characteristics of IDRs supporting clinical research at a selection of major academic health centers.
General status and data sources
Based on responses to the 2007 survey, an IDR was defined as ‘a clinical data repository that utilizes a non-transactional structure optimized for research purposes rather than clinical care.’ In addition, six optional qualifiers were identified:
Contains clinical, administrative, trial, and omics data
Incorporates common data standards across different databases
Includes a data quality component for continuous improvement based on feedback from end users
Includes a metadata repository function with information on data quality (eg, rates of missing values)
Integrates with institutional review board (IRB) systems for streamlined access to data for research purposes
Structured to provide statistical support so that high quality clinical and translational research can result from analysis.
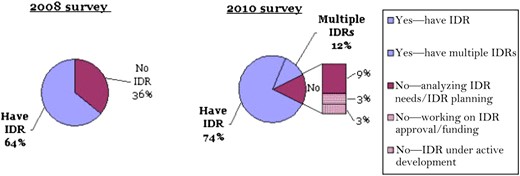
Based on this definition, 18 of 28 institutions (64%) reported having an IDR in 2008. By 2010, 30 of 35 (86%) indicated their organization had one or more clinical data repositories. Seventeen of 35 (48%) considered their IDR to be a dedicated research data repository. Thirteen of 35 overall (37%) contained data from multiple institutional entities, with five of those 13 designated for research only. Four of 35 (11%) reported having multiple repositories (figure 1). Of those four, only one institution described the multiple IDRs as designated for research only. Responses of rough estimates of patient populations (22 of 35) varied widely, from ‘43,000 unique patients' to 10 million. Discarding the highest and lowest values as outliers resulted in a mean of 1.8 million patients, median 1.6 million. Data repository sizes reported (25 of 35) also ranged broadly from 10 GB to over 100 TB, with a mean of 2.4 TB and median of 1.3 TB, again excluding the highest and lowest values. This question was not asked in 2008. Responses for both of these questions were collected as free text in order to enable descriptive qualifications and to avoid assumptions that would be required to designate pre-set ranges.
Does your organization have an integrated data repository (IDR)?
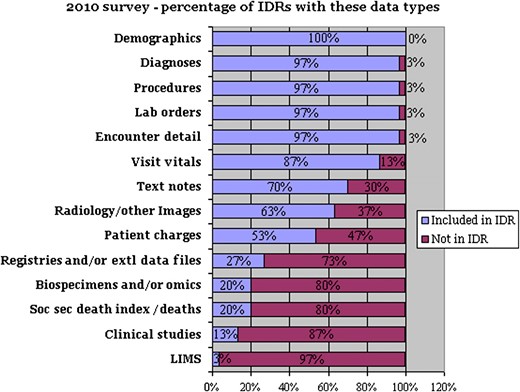
Both the 2008 and 2010 surveys sought to identify specific types of clinical data (eg, demographics, laboratory tests, medications), and duration of history for each of these data types (from 0 to over 10 years). Responses suggest that over time, data included have shifted from primarily administrative to incorporating more of the data contained in source electronic medical records and supporting systems (figure 2). In addition, a number of IDRs are including research-focused data, such as genomic and proteomic data (20%) or clinical-trial specific data (13%).
Percentage of integrated data repositories (IDRs) that incorporate these data areas. Extl, external; LIMS, laboratory information management system; Soc sec, social security.
Technology selection and staffing
Between the 2008 and 2010 surveys, use of Oracle as a database management system to support IDRs increased from 47% to 60% of the organizations, and the use of SQL Server increased from 18% to 47%. Use of ‘in-house’ developed front-facing business intelligence tools decreased from 71% to 40%, while adoption of the open-source data warehouse tool i2b2 (Informatics for Integrating Biology and the Bedside)6 increased from 12% to 30%. In 2008, the average number of support staff for a data repository was 7.7 full-time equivalents (FTEs) compared with an average of 6.42 FTEs in 2010. Respondents were also asked about clinical systems in use at their institutions, broken down by inpatient, outpatient, radiology, medications, and emergency department. This question was asked regardless of whether the institution had an IDR or used a clinical system as an IDR data source. Cerner and Epic continue to be represented strongly, falling within the top three system vendors across multiple clinical system areas (see appendix B for details).
Data access and delivery
A significant change was seen in how IDR functions support researchers' access to clinical data. Between the 2008 and 2010 surveys, researcher ‘self-serve’ access rose in de-identified aggregate patient data (21% in 2008 vs 54% in 2010), de-identified patient-level data (14% vs 31%), and identifiable patient data (8% vs 25%). At the same time, the percentage of institutions not providing datasets dropped for all three categories (table 2).
How do your researchers access your data?
| . | Receive result set from intermediary . | Self-service query access . | Not provided . | Other . | Responses . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Access option . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . |
| De-identified aggregate patient data | 32% (9) | 40% (12) | 21% (6) | 54% (16) | 21% (6) | 3% (1) | 25% (7) | 3% (1) | 28 | 30 |
| De-identified patient-level data | 39% (11) | 63% (18) | 14% (4) | 31% (9) | 10% (3) | 3% (1) | 35% (10) | 3% (1) | 28 | 29 |
| Fully identified patient-level data | 44% (11) | 71% (20) | 8% (2) | 25% (7) | 20% (5) | 0 | 28% (7) | 4% (1) | 25 | 28 |
| . | Receive result set from intermediary . | Self-service query access . | Not provided . | Other . | Responses . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Access option . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . |
| De-identified aggregate patient data | 32% (9) | 40% (12) | 21% (6) | 54% (16) | 21% (6) | 3% (1) | 25% (7) | 3% (1) | 28 | 30 |
| De-identified patient-level data | 39% (11) | 63% (18) | 14% (4) | 31% (9) | 10% (3) | 3% (1) | 35% (10) | 3% (1) | 28 | 29 |
| Fully identified patient-level data | 44% (11) | 71% (20) | 8% (2) | 25% (7) | 20% (5) | 0 | 28% (7) | 4% (1) | 25 | 28 |
How do your researchers access your data?
| . | Receive result set from intermediary . | Self-service query access . | Not provided . | Other . | Responses . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Access option . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . |
| De-identified aggregate patient data | 32% (9) | 40% (12) | 21% (6) | 54% (16) | 21% (6) | 3% (1) | 25% (7) | 3% (1) | 28 | 30 |
| De-identified patient-level data | 39% (11) | 63% (18) | 14% (4) | 31% (9) | 10% (3) | 3% (1) | 35% (10) | 3% (1) | 28 | 29 |
| Fully identified patient-level data | 44% (11) | 71% (20) | 8% (2) | 25% (7) | 20% (5) | 0 | 28% (7) | 4% (1) | 25 | 28 |
| . | Receive result set from intermediary . | Self-service query access . | Not provided . | Other . | Responses . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Access option . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . | 2008 . | 2010 . |
| De-identified aggregate patient data | 32% (9) | 40% (12) | 21% (6) | 54% (16) | 21% (6) | 3% (1) | 25% (7) | 3% (1) | 28 | 30 |
| De-identified patient-level data | 39% (11) | 63% (18) | 14% (4) | 31% (9) | 10% (3) | 3% (1) | 35% (10) | 3% (1) | 28 | 29 |
| Fully identified patient-level data | 44% (11) | 71% (20) | 8% (2) | 25% (7) | 20% (5) | 0 | 28% (7) | 4% (1) | 25 | 28 |
As CTSA organizations have built or expanded their data access capabilities, they have also increased their use of de-identification capabilities. In the 2010 IDR survey, 55% (16 of 29) of organizations indicated they are utilizing automated de-identification of patient-level data (either while data is loaded to the repository or during data access). Of the organizations with clinical text notes in their IDRs, 33% (7 of 21) indicated they include text clinical notes in their automated de-identification processes.
Project experiences
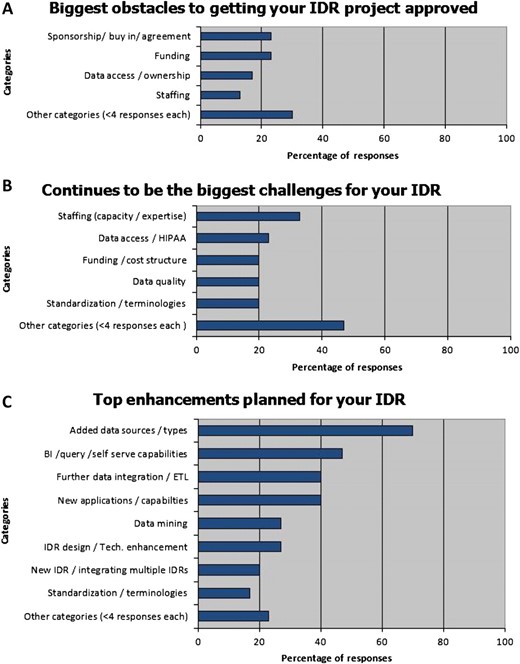
The survey asked respondents to identify challenges experienced in deploying and maintaining these systems. Funding and gaining sponsorship/stakeholder buy-in were the top reported development obstacles (23%), followed by data ownership and access issues (17%), and staffing (13%). Continuing challenges were staffing (33%), data access/Health Insurance Portability and Accountability Act (HIPAA) issues (23%), standardization/terminologies (20%), and data quality issues (20%). Top reported future enhancements were adding new data sources (70%) followed by business intelligence/query/self-serve capabilities (47%) (see figure 3).
Obstacles, challenges, and enhancements to integrated data repository (IDR) development. BI, business intelligence; HIPAA, Health Insurance Portability and Accountability Act.
Discussion
These results highlight the growing range of approaches and capabilities of IDRs in meeting CTSA organizations' research needs for clinical data, and provide a current snapshot of the variation in CTSA IDRs in terms of size, scope, access modes, maturity of current and anticipated data types being incorporated, and expectations of use. These results, although derived from a select subset of academic health centers, will be useful for consideration by prospective implementers of research IDRs in other settings as they reflect challenges, solutions, and trends for adopting and sustaining such systems.
Several key changes from the 2008 survey data to the present were observed. There appears to be increasing movement away from ‘home-grown’ solutions to more commonly used system platforms such as i2b2. The increasing percentage of institutions that report use of an active master patient index (up 23% over 2008) also suggests a trend toward the use of centralized IDRs, whether vendor based solutions or open source, rather than ad hoc collections of data. As organizations expand their use of EHRs, the data available in IDRs is transitioning from primarily administrative to inclusive of core clinical data elements. A small number of respondents indicated recent inclusion of omics data, predictably reflecting an increasing trend toward genomic and personalized medicine.7
Increasingly, IDR administrators are providing a range of web-based, self-serve access modes to these repositories, for both de-identified and patient identifiable data. One unexpected finding was the fact that the average number of reported FTEs has decreased. This might reflect the need for significant up-front development efforts, with decreased resources required for long-term maintenance, increased automation in data access, a depressed funding environment, or some combination of these factors.
With regard to the goal aimed at identifying areas for future exploration, these results have suggested, or helped refine, a number of new avenues for inquiry. For example, we wish to capture information on organizational readiness for optimal use of EHR data, whether internally or for larger scale data sharing, and issues encountered by organizations with multi-institution data repositories. The fact that more than one third of institutions with IDRs report that their system contains data from multiple institutions suggests that inter-institutional governance issues are beginning to be successfully addressed. A key component of these assessments will be to ascertain which institutions have implemented appropriate access and use policies that can support increased use of these resources, including approaches to de-identification, and how they have accomplished this. We plan to explore whether such an increase in data access can be used as a measurement of impact on translational research efforts. Another area for further exploration that had not been anticipated is the issue of how to effectively capture and share information about implementations of research data repositories. Ongoing analysis in this regard will enable us to refine our approach in the development of a new IDRs survey to be deployed in 2012.
Finally, a key and recurring question is how to best sustain the deployment of these resources in a constrained funding climate. We seek to identify the extent to which sites are attempting to reduce costs by leveraging open-source and consortia software development, as opposed to selecting vendor-based solutions. We would also like to explore how institutions have leveraged functionality designed primarily to serve the clinical care enterprise, and therefore paid for by their respective hospitals and medical centers, to facilitate secondary use for research as well.
Limitations
Given the length of the survey (77 questions), we chose to minimize explanations and keep questions succinct to encourage completion of the full survey. However, based on our analysis and on specific inquiries from participating institutions, it is apparent that some of the questions were open to interpretation. For example, answers to the questions regarding how long specific types of data had been included, together with answers regarding how long the IDR had been available for research, suggest that some interpreted the questions as being about the number of years researchers had access to the different types of data, while others interpreted it as asking from how many years back the available data were derived. These questions will be revisited in the next survey. In addition, of the 35 individuals who responded in 2010, only 12 of those had paired controls in that they were the same individuals who had responded in 2008. Some portion of the differences observed may have come from differences in subjective interpretation, or in perspectives from respondents. Some quantitative questions were asked in free text form in order to obtain estimates unbiased by pre-defined values or ranges. Given the empirical range of responses from this version, future surveys will be able to capture these data in a more structured way.
Although the CTSA consortium provides a convenient infrastructure through which to carry out a project like ours, it does represent a specific, and in some respects biased, subset of large health centers nationally. In the future, we would like to expand the scope of our inquiry through partnerships with organizations such as the Healthcare Data Warehousing Association4 and The Data Warehousing Institute.8
While the underlying IDR information technology is of interest, the survey is also meant to target data availability, access, and use. These distinct facets will be addressed more explicitly in future surveys. Finally, we did not explicitly ask for respondents' roles or the characteristics of individual CTSAs that would be useful in cross-site comparison, such as relationship with an academic medical center, or availability of centralized research IT resources.
Conclusion
The survey responses described here provide insights into the adoption and use of IDRs by a number of academic medical centers, as well as emerging trends in this rapidly evolving arena. The results have already proven to be of value to members of our working group with specific questions such as how other institutions allow non-aggregated, patient-level access to identified data, including protected health information. We see the survey results as providing an evolving map of characteristics and issues encountered in the large-scale development of IDRs in the USA. As a resource, we hope that it will provide a basis for further research in clinical research informatics, as well as identification of opportunities for enhanced resource sharing and collaboration.
Funding
This project has been funded in whole or in part with Federal funds from the National Center for Research Resources, National Institutes of Health (NIH), through the Clinical and Translational Science Awards Program (CTSA), part of the Roadmap Initiative, Re-Engineering the Clinical Research Enterprise. The manuscript was approved by the CTSA Consortium Publications Committee. Authors were supported by grants through the NIH National Center for Research Resources, Duke Translational Medicine Institute (UL1 RR024128), Oregon Clinical Translational Research Institute (UL1 RR024140), Scripps Translational Science Institute (UL1 RR025074), University of Alabama at Birmingham Center for Clinical and Translational Science (UL1 RR025777), and the Institute of Translational Health Sciences, University of Washington (UL1 RR025014).
Competing interests
None.
Provenance and peer review
Not commissioned; externally peer reviewed.
References