- 1State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
- 2State Key Laboratory of Crop Genetics and Germplasm Enhancement, MOA Key Laboratory of Plant Nutrition and Fertilization in Lower-Middle Reaches of the Yangtze River, Nanjing Agricultural University, Nanjing, China
Salinity imposes a major constraint over the productivity of rice. A set of chromosome segment substitution lines (CSSLs), derived from a cross between the japonica type cultivar (cv.) Nipponbare (salinity sensitive) and the indica type cv. 9311 (moderately tolerant), was scored using a hydroponics system for their salinity tolerance at the seedling stage. Two of the CSSLs, which share a ∼1.2 Mbp stretch of chromosome 4 derived from cv. Nipponbare, were as sensitive to the stress as cv. Nipponbare itself. Fine mapping based on an F2 population bred from a backcross between one of these CSSLs and cv. 9311 narrowed this region to 95 Kbp, within which only one gene (OsHAK1) exhibited a differential (lower) transcript abundance in cv. Nipponbare and the two CSSLs compared to in cv. 9311. The gene was up-regulated by exposure to salinity stress both in the root and the shoot, while a knockout mutant proved to be more salinity sensitive than its wild type with respect to its growth at both the vegetative and reproductive stages. Seedlings over-expressing OsHAK1 were more tolerant than wild type, displaying a superior photosynthetic rate, a higher leaf chlorophyll content, an enhanced accumulation of proline and a reduced level of lipid peroxidation. At the transcriptome level, the over-expression of OsHAK1 stimulated a number of stress-responsive genes as well as four genes known to be involved in Na+ homeostasis and the salinity response (OsHKT1;5, OsSOS1, OsLti6a and OsLti6b). When the stress was applied at booting through to maturity, the OsHAK1 over-expressors out-yielded wild type by 25%, and no negative pleiotropic effects were expressed in plants gown under non-saline conditions. The level of expression of OsHAK1 was correlated with Na+/K+ homeostasis, which implies that the gene should be explored a target for molecular approaches to the improvement of salinity tolerance in rice.
Introduction
Salinity imposes a serious constraint over crop productivity, especially for crops grown under irrigation or in coastal lowlands prone to seawater ingress (Campbell et al., 2017). Over 800 Mha of arable land is affected by salinity to some degree, an area which represents some 6% of the total used for cropping (Munns and Tester, 2008). While certain cereal species show a substantial level of tolerance to salinity, rice does not (Munns and Tester, 2008). Its sensitivity depends on the developmental stage of the plant (Fageria, 1985; Khan et al., 1997); it is particularly vulnerable during its early vegetative growth, so that stand density is compromised in salinity-affected fields (Lutts et al., 1995; Gregorio et al., 2002). Some cultivars exhibit their highest sensitivity at the time of tillering and panicle initiation (Zeng et al., 2002).
Surveys of rice germplasm have suggested the presence of considerable genetic variation for the ability to withstand salinity (Negrão et al., 2011): the most tolerant entries known are originate from coastal environments (Flowers and Yeo, 1995; Gregorio et al., 2002; Flowers, 2004; Thomson et al., 2010). The genetic control of salinity tolerance, revealed using the quantitative trait locus approach (Koyama et al., 2001; Lin et al., 2004; Thomson et al., 2010; Ahmadi et al., 2011; Negrão et al., 2011; Platten et al., 2013), involves genes present in a number of genomic regions. The best characterized of these forms part of the short arm of chromosome 1, the site of HKT1;5 (Lin et al., 2004; Thomson et al., 2010; Ahmadi et al., 2011), a gene which has been shown to encode a sodium ion (Na+) transporter which regulates shoot Na+/K+ homeostasis during an episode of salinity stress (Ren et al., 2005).
The potassium cation (K+) is involved in many aspects of plant growth and development (Ahmad et al., 2016). In saline soils, its uptake across the plasma membrane is impeded by competition with Na+, resulting in a fall in the plant tissue’s [K+]/[Na+] ratio (Schachtman and Liu, 1999). Plants have evolved a number of means of both promoting the uptake and transport of K+ and limiting the accumulation of Na+ in sensitive tissues (Schachtman and Liu, 1999; Luan et al., 2009; Cui et al., 2016). The transporters encoded by KT/HAK/KUP genes are thought to be key for maintaining an acceptable [K+]/[Na+] ratio, and hence represent an important component of plant tolerance to salinity stress (Chen et al., 2015b). A number of KT/HAK/KUP genes have been shown to be inducible by salinity stress (Maathuis, 2006). Both HvHAK1 (Santa-María et al., 1997) and PhaHAK1 (Takahashi et al., 2007) are likely involved in low affinity Na+ influx. A single altered residue in the HAK5 protein produced by several plant species has the effect of raising both the protein’s affinity for K+ and the level of salinity tolerance exhibited by yeast cells expressing this protein heterologously (Alemán et al., 2014). The rice OsHAK2 protein was sensitive to extracellular Na+ and transports Na+ more effectively than K+ (Horie et al., 2011b). The expression of OsHAK5 in tobacco BY2 cells enhances their salinity tolerance via a boost in their capacity to accumulate K+ without affecting Na+ uptake (Horie et al., 2011b). The over-expression of OsHAK5 in rice increases the shoot [K+]/[Na+] ratio and enhances the level of its salinity stress tolerance (Yang et al., 2014), while the knocking out of OsHAK1 retards growth and depresses K+ accumulation (Chen et al., 2015b).
Here, a set of rice chromosome segment substitution lines (CSSLs), developed from a cross between the cultivars Nipponbare and 9311, was used to characterize variation for salinity tolerance at the seedling stage and to identify the genetic basis of K+ and Na+ homeostasis. The main outcome of the experiments was a demonstration that the level of OsHAK1 expression has an impact on the plant’s ability to tolerate salinity, suggesting the gene could be targeted in breeding programs seeking to boost the salinity tolerance of rice.
Materials and Methods
Plant Materials and Growing Conditions
From a new set of CSSLs developed from the cross between the japonica type cv. Nipponbare (donor) and the indica type cv. 9311 (recipient), the two lines CSSL-1 and CSSL-2 were chosen to analyze the genetic basis of the differential salinity response of cv. 9311 and cv. Nipponbare. Both CSSLs were poor performers when challenged by salinity stress. To create a mapping population, CSSL-1 was back-crossed with cv. 9311 and the F1 plants allowed to self-pollinate. The origin and properties of two transgenic lines, one harboring pOsHAK1::GUS and the other pOsHAK1::OsHAK1, along with the oshak1 mutant in a background of either cv. Dongjin or cv. Manan, have been described by Chen et al. (2015b). To raise seedlings under hydroponics, the grains were surface-sterilized and allowed to germinate in a nutrient-containing solution (Li et al., 2006); after 1 week, uniformly sized seedlings were transferred to a hydroponics system and exposed to a 14 h photoperiod, under a temperature regime of 30°C/22°C and a relative humidity level of ∼70%. The nutrient solution was replaced every 2 days. After a further week, the solution was salinized by the addition of 100 mM NaCl, to which the seedlings were exposed for a week. The harvested material was immersed in 0.1 mM CaSO4 for 5 min, then separated into root and shoot, which were weighed separately and assayed for their [Na+] (Na+ content) and [K+], following methods given by Chen et al. (2018). For a soil-based experiment set up to compare the performance of the OsHAK1 transgenic and wild type (WT) plants, single plants were grown in 10 kg of air-dried loam for 5 weeks, then exposed to a 3 week period of salinity stress (1.5 g NaCl per kg of soil) by following the Zeng et al. (2017) procedure; a duplicate set of plants, to which no salt was provided, was grown as a control. A second experiment applied the same level of stress to plants once they had reached the booting stage, with the stress maintained through to plant maturity. Each treatment was replicated five times.
Quantitative Real Time PCR (qRT-PCR)
The procedures used to conduct qRT-PCR followed those specified by Chen et al. (2015a). RNA was extracted from root and shoot tissue of both WT and transgenic, hydroponically grown seedlings, and from the youngest two leaves of soil-grown plants. The rice Ubq gene (LOC_Os03g13170) was chosen as the reference sequence, and relative transcript abundances were calculated following the suggestion of Li Y. et al. (2014). The stress-responsive genes assayed were OsP5CS1 (pyrroline-5-carboxylate synthetase; Sripinyowanich et al., 2013), OsDREB2A (dehydration-responsive element-binding protein; Dubouzet et al., 2003), OsAP37 (adaptor protein; Oh et al., 2009), and OsERD1 (early responsive to dehydration; a homolog of Arabidopsis thaliana AtERD1; Kiyosue et al., 1994), while the transporter genes were OsHKT1;5 (Na+ transporter; Ren et al., 2005), OsSOS1 (Na+/H+ antiporter; Martínez-Atienza et al., 2007), OsLti6a and OsLti6b (proteins involved in maintaining the integrity of the plasma membrane during abiotic stress; Mekawy et al., 2015). The sequences of the various primers used are listed in Supplementary Table 1.
Quantification of GUS Activity
GUS activity was measured in transgenic plants harboring the pOsHAK1::GUS construct as described by Chen et al. (2015a).
Functional Complementation of OsHAK1 in Yeast
The coding sequence of OsHAK1 was inserted into pYES2 (Invitrogen, Carlsbad, CA, United States) driven by the inducible Gal1 promoter, then transformed into the salinity-sensitive yeast strain AXT3 [Δena1::HIS3::ena4,nha1::LEU2,Δnhx1::TRP1] (Li Q. et al., 2014). The cells were grown for 3 days on solidified AP medium at 30°C (Horie et al., 2011a). Their ion content was measured in three independent clones per construct, using the Xu et al. (2008) method, with the following modifications: the cells were cultured at 30°C in 2 L liquid AP medium either with or without 50 mM NaCl with shaking (200 rpm). When the OD600 reached 0.8, they were harvested by centrifugation (5 min, 8000 rpm) and rinsed twice in liquid AP medium with no added K+ or Na+. They were then dried at 70°C for 72 h, weighed and digested overnight in 1 mL HNO3. After diluting with de-ionized water, the content of both K+ and Na+ was measured using an Optima 2100DV ICP-emission spectrometer (PerkinElmer, Waltham, MA, United States).
Quantification of Electrolyte Leakage
The methods used to quantify electrolyte leakage were as described by Chen et al. (2017).
Measurement of Photosynthetic Rate
Seedling rates of photosynthesis were measured between 9.00 and 11.00 am using a Li-COR6400 portable photosynthesis system equipped with a LED leaf cuvette (Li-COR, Lincoln, NE, United States), as described by Li et al. (2016). At least five individuals per genotype per each stress treatment were assayed.
Determination of Leaf Chlorophyll Content
Following the procedures given by Li et al. (2016), leaves of WT and transgenic plants were weighed and extracted in aqueous 95% (v/v) ethanol. The absorbance of the extract was recorded at both 663 nm and 645 nm using a UV2400 spectrophotometer (Shimadzu, Kyoto, Japan). Chlorophyll concentrations (mg per g leaf fresh weight) were derived from the expression 8.02A663 + 20.21A645.
Leaf Hydrogen Peroxide (H2O2), Malondialdehyde (MDA) and Proline Content
The H2O2 content of leaf samples was determined following their equilibration in 5.3 mM TiCl4 dissolved in 3.7 M H2SO4, based on the Mostofa and Fujita (2013) procedure. Leaf MDA contents were assessed following Chen et al. (2017), and leaf proline contents following Bates et al. (1973).
Statistical Analysis
Analyses of variance were carried out using routines implemented in SPSS v10 software (SPSS Inc., Chicago, IL, United States). Statistically significant (P < 0.05) differences between the performance either of the various genotypes and WT or between different treatments (as determined by Tukey’s test), are denoted by the inclusion on the histograms of a different letter or an asterisk.
Results
The Relative Salinity Tolerance of the CSSLs and Their Parental Cultivars
At the seedling stage, cv. 9311 was more tolerant of a 7 days exposure to 100 mM NaCl than was cv. Nipponbare. The cv. Nipponbare seedlings displayed both extensive wilting and foliar chlorosis, whereas the cv. 9311 ones became only mildly wilted, displaying withering just at the tip of their leaf blades (Figures 1A,B). Under control conditions, cv. 9311 seedlings accumulated the most root and shoot dry matter, followed by those of CSSL-1, CSSL-2 and cv. Nipponbare. The effect of the salinity stress was to reduce the dry matter accumulated by cv. 9311 by 29%, that by CSSL-1 by 34%, that by CSSL-2 by 31% and that by cv. Nipponbare by 36% (Figure 1C). Measurement of the root and shoot [K+] and [Na+] in seedlings grown under salinity stress showed that the parental cultivars differed significantly from one another: [K+] was 10% greater in the root and 15% greater in the shoot of stressed cv. 9311 seedlings than of cv. Nipponbare seedlings (Figure 1D). The effect of the stress on the two CSSLs was to raise the root and shoot [K+] above the level induced in the cv. Nipponbare seedlings, but below that induced in cv. 9311 seedlings. The root and shoot [K+]/[Na+] ratios behaved in a similar fashion (Figure 1E).
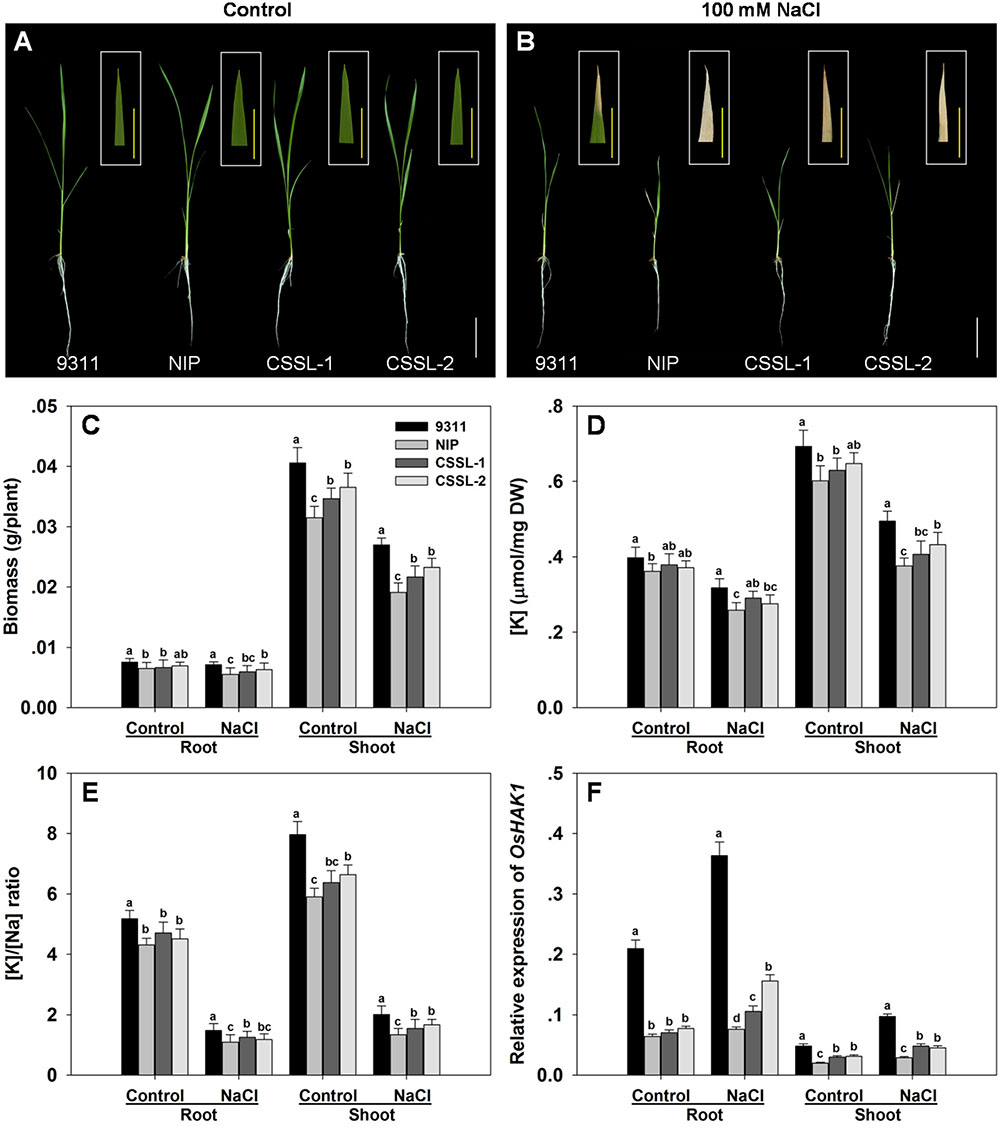
FIGURE 1. The contrasting salinity tolerance of the rice cvs Nipponbare and 9311, and two derived CSSLs. Seedlings were exposed to 100 mM NaCl for 7 days. (A,B) The growth and appearance of the plants raised in (A) the absence of NaCl, (B) the presence of 100 mM NaCl. White bars: 5 cm, yellow bars: 1 cm. (C) Dry matter accumulation, (D) [K+], (E) [K+]/[Na+] ratio and (F) the abundance of OsHAK1 transcript. Values shown in the form of mean ± SE (n = 5). Means labeled with the same lower case letter did not differ significantly from one another (P < 0.05).
The CSSL-1 and CSSL-2 genomes harbor a non-identical spectrum of cv. Nipponbare segments, but they share an overlap on chromosome 4 defined by the two InDel markers IND4-3 and IND4-13. The 1,774 F2 segregants derived from the cross CSSL-1 × cv. 9311 were tested with respect to their seedling response to salinity and also genotyped with respect to IND4-3 and IND4-13, along with a further four newly developed InDel markers lying within the chromosome 4 segment (Supplementary Table 2). Combining the genotypic and phenotypic data reduced the genomic window harboring gene(s) involved in the differential salinity response of cvs Nipponbare and 9311 to a 95 Kbp segment bounded by IND4-9 and IND4-10 (Figure 2), a region which, according to the Rice Genome Annotation Project1, contains 11 annotated genes. One of these genes is OsHAK1, which encodes a high affinity potassium transporter important for the regulation of K+ homeostasis (Supplementary Table 3). When the transcription in both the root and shoot of all eleven annotated genes was assessed, it was clear that the abundance of OsHAK1 transcript was different between cv. Nipponbare and both CSSLs compared to cv. 9311 (Figure 1F), while there was no evidence for differential transcription among any of the other ten genes (data not shown). A sequence analysis of the entire OsHAK1 coding sequence revealed a single synonymous nucleotide substitution between cvs Nipponbare and 9311 (Supplementary Figure 1).
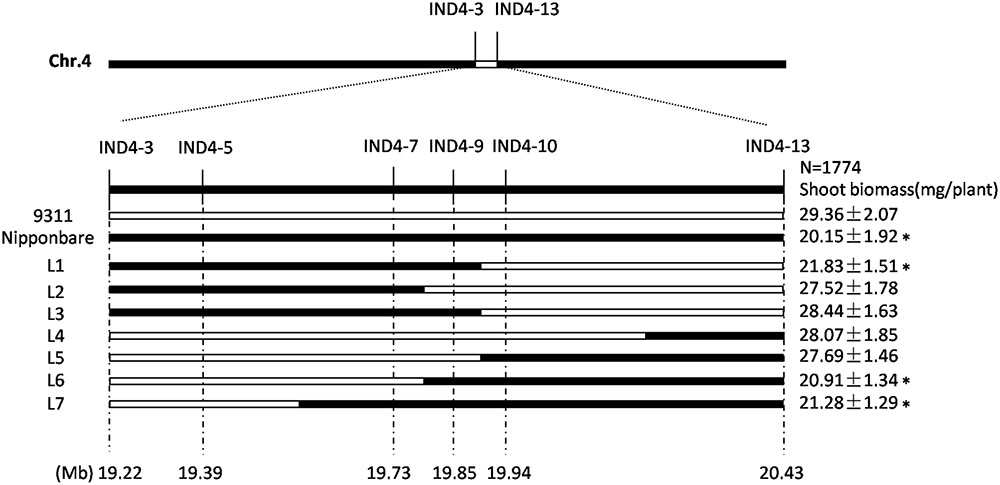
FIGURE 2. Graphical genotypes uncovered in the critical region of chromosome 4 among F2 progeny derived from the cross CSSL-1 x cv. 9311. The unfilled regions represent those homozygous for cv. 9311 alleles, and the filled ones represent those homozygous for cv. Nipponbare alleles. Mean shoot biomass values ± SE (n = 5) are shown on the right. ∗Mean differs significantly (P < 0.05) between cv. 9311 and genotypes carrying the critical chromosome 4 segment inherited from cv. Nipponbare.
The Response of OsHAK1 to Salinity Stress
A qRT-PCR analysis showed that OsHAK1 was induced in both root and shoot – but particularly in the former - by exposure of the seedlings to 100 mM NaCl (Figure 3). In the roots, the abundance of transcript rose within 1 h of the imposition of the stress, peaked to a level equivalent to three times the background within 3 h, then gradually declined over the rest of the assessment period (Figure 3A). In the shoots, the abundance of transcript was highest after 6 h of exposure (double the background level) (Figure 3B). OsHAK1 expression was also examined in the roots and leaf blades of plants by analyzing GUS activity in plants carrying the pOsHAK1::GUS transgene (Supplementary Figures 2A–D). The transgene’s activity was significantly boosted by exposure to 100 mM NaCl, and more so in the root than in the shoot (Supplementary Figure 2E), consistent with the transcriptomic analysis (Figure 3).
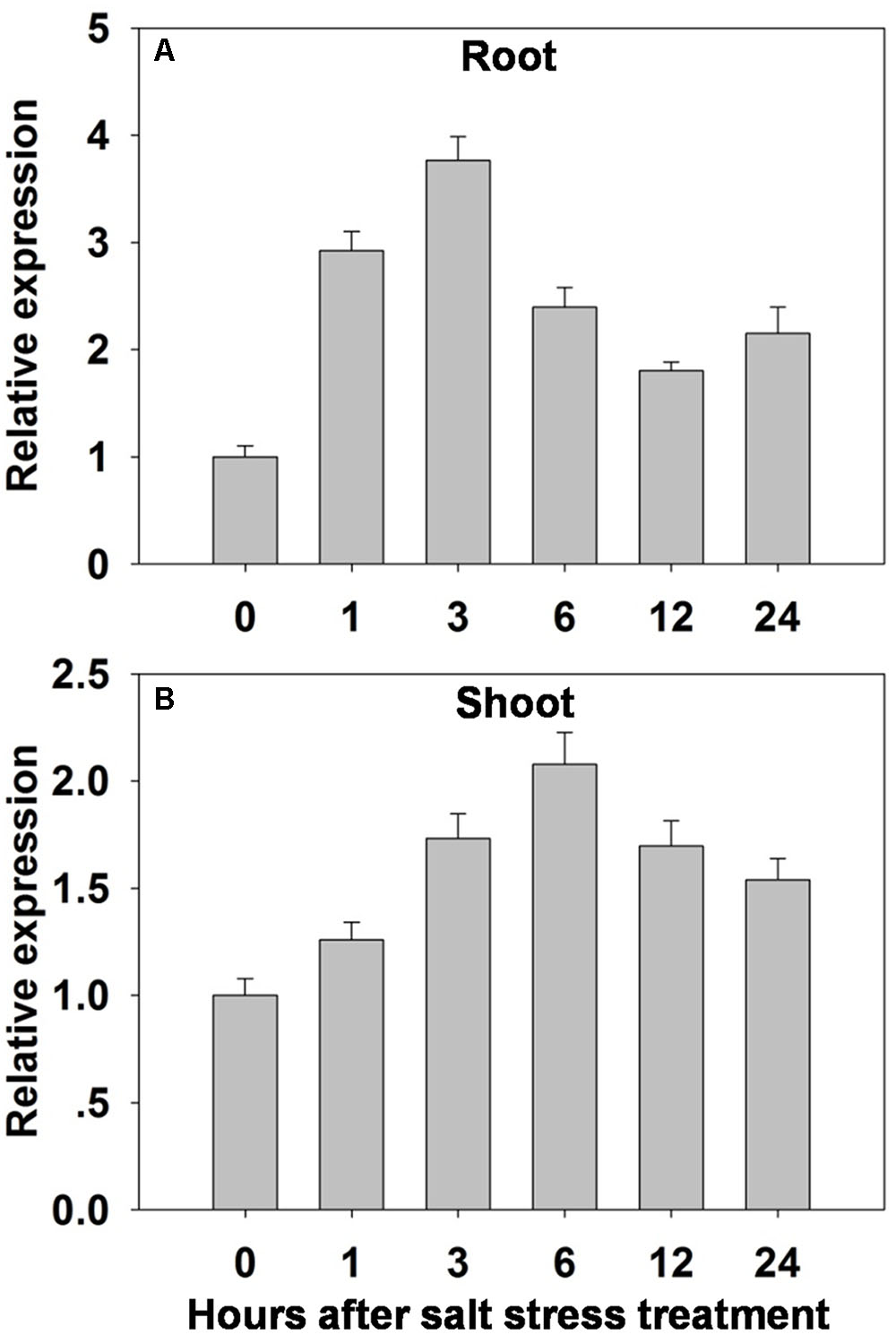
FIGURE 3. The effect of salinity stress (100 mM NaCl for 0–24 h) on the transcription of OsHAK1 in (A) the roots and (B) the shoots of WT seedlings. Transcript abundances were derived using qRT-PCR, with Ubq chosen as the reference sequence. The abundance of OsHAK1 transcript prior to the salinity treatment was set arbitrarily as 1. The whiskers indicate the SE (n = 3).
The Functionality of OsHAK1 in a Yeast Mutant Lacking Na+/H+ Antiporter Activity
Cells of yeast strain AXT3 transformed with OsHAK1 grew better on AP medium containing either 50 mM or 100 mM NaCl than did those carrying just an empty pYES2 vector (Figure 4A). Measurement of their [K+] and [Na+] showed that in the absence of salinity stress, the control and transgenic cells were indistinguishable (Figures 4B,C). Exposure to the stress raised [Na+] and decreased [K+] in both control and transgenic cells. However, [Na+] in the OsHAK1 expressors was markedly lower than in the non-expressors (Figure 4C), while [K+] was 43% greater (Figure 4B).
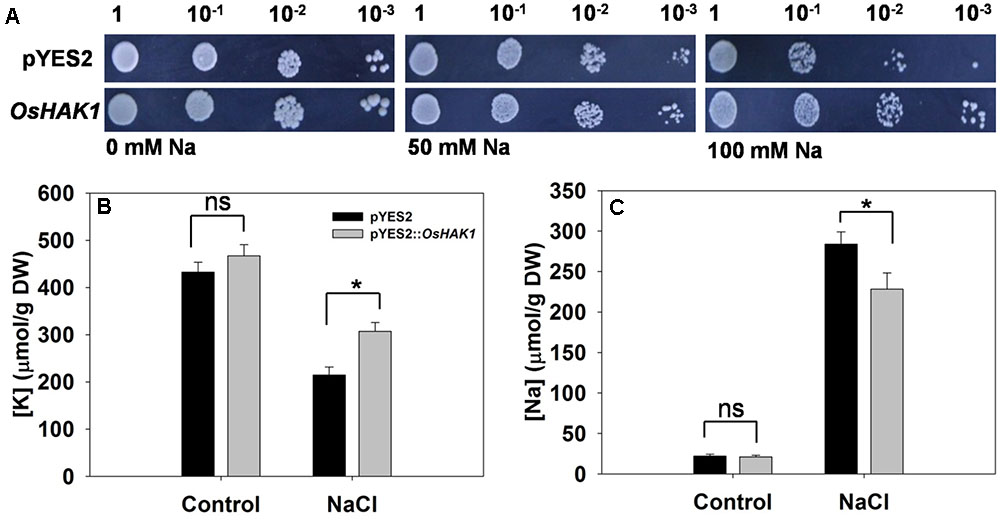
FIGURE 4. Complementation of a salinity-sensitive yeast mutant by OsHAK1. (A) Yeast cells harboring an empty vector (pYES2) are shown in the upper row, and those harboring OsHAK1 in the lower row. The (B) [K+] and (C) [Na+] of yeast cells grown either in the absence (control) or presence of 50 mM NaCl. Data represent the mean ± SE (n = 3). Significant differences (P < 0.05) between the performance of cells harboring the empty vector and those harboring the OsHAK1 transgene are indicated by an asterisk. ns, non-significant difference; DW, dry weight.
The Effect on Seedling Growth of Altering the Level of OsHAK1 Transcript
The response of hydroponically grown seedlings to exposure to 100 mM NaCl was compared between WT, oshak1 knock-out mutant and OsHAK1 over-expressing plants. No differences were detected when the plants were grown in the absence of salinity (Figures 5A,B), but the imposition of stress induced wilting and foliar chlorosis in WT and oshak1, but not in the OsHAK1 over-expressor (Figures 5A,B). The stress did suppress shoot and root growth for all three genotypes, but the extent of the reduction was significantly less for the OsHAK1 over-expressor than for the knock-out mutant plants. When challenged by salinity, the plant height, root dry matter and shoot dry matter attained by the over-expressor seedlings were all significantly greater than those attained by WT seedlings. The total dry matter accumulated by the knock-out mutants was about 85% of that accumulated by their respective WT (Figures 5C,D).
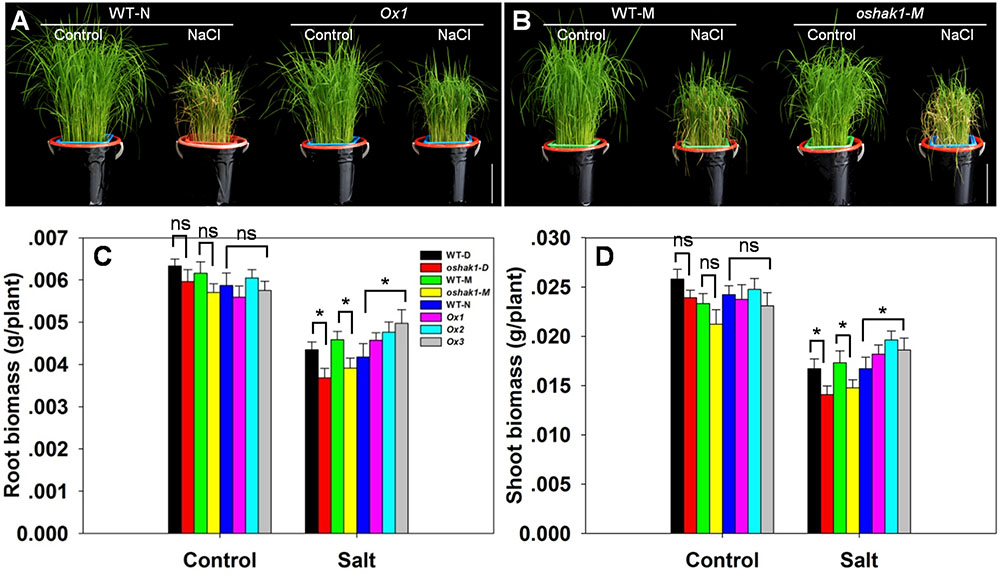
FIGURE 5. The effect of OsHAK1 on the growth of rice seedlings. (A) The growth of an OsHAK1 over-expressor (Ox1) and WT in either non-salinized (control) or salinized (100 mM NaCl) medium. (B) The growth of an OsHAK1 knock-out mutant (oshak1-M) and its WT in either non-salinized (control) or salinized (100 mM NaCl) medium. Bar: 10 cm. (C,D) Dry matter accumulation of seedings raised in either non-salinized (control) or salinized (100 mM NaCl) medium: (C) the root and (D) the shoot. Values shown in the form mean ± SE (n = 5). Significant differences (P < 0.05) between the test genotype and WT are indicated by an asterisk. ns, non-significant difference; WT-D, cv. Dongjin; WT-N, cv. Nipponbare; WT-M, cv. Manan, oshak1-D, oshak1-M: OsHAK1 knockout mutants in, respectively a cv. Dongjin and a cv. Manan background.
The Effect of OsHAK1 on Ion Uptake and the Transcription of Other Transporter Genes
There was no significant variation between the genotypes with respect to either [Na+] or the [Na+]/[K+] ratio in either root or shoot under control conditions; however, in the presence of 100 mM NaCl, compared to the performance of WT plants, the oshak1 mutants over-accumulated Na+, while the OsHAK1 over-expressors took up significantly less Na+ into both their roots and shoots (Figures 6A,B). The [Na+]/[K+] ratio was particularly high for the mutant seedlings, while it was maintained at a lower level in the over-expressors (Figures 6C,D). The qRT-PCR platform was deployed to quantify the transcription of four genes encoding a variety of transport proteins. OsHKT1;5 was significantly down-regulated in the oshak1 mutant plants under control conditions (Figure 7A). The presence of salinity down-regulated OsHKT1;5 in each of the genotypes, although the extent of the reduction was less for the OsHAK1 over-expressor than for the knock-out mutant plants (Figure 7A). For the other three genes, under control conditions, there was no variation in the level of their transcription between the various lines, but under salinity stress the genes were all up-regulated in the over-expressor relative to WT and down-regulated in the knock-out mutants (Figures 7B,D).
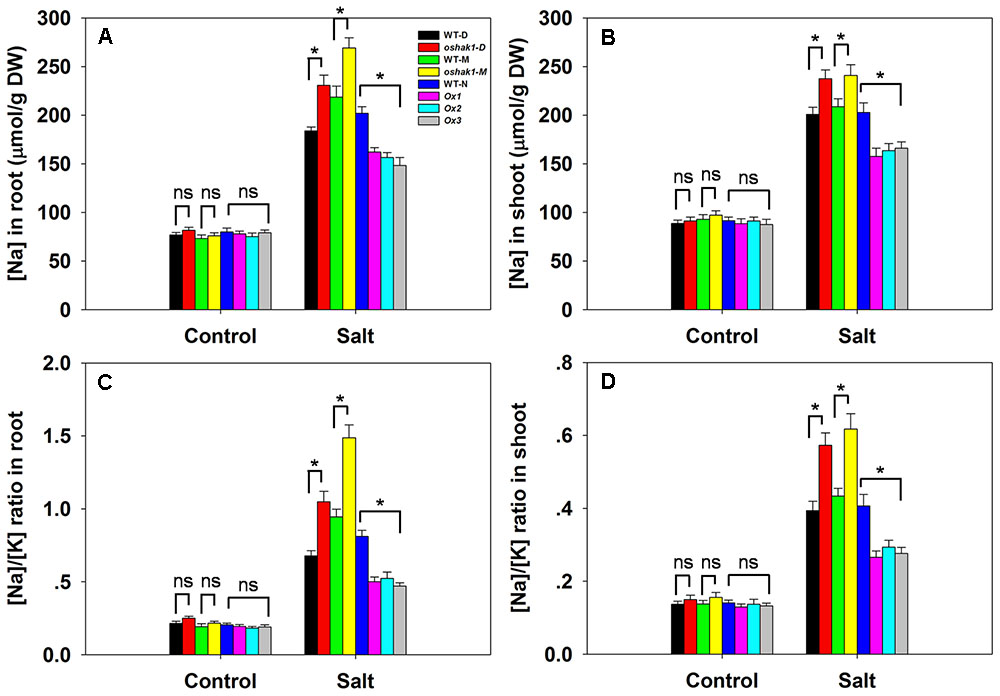
FIGURE 6. The effect of OsHAK1 on Na+ accumulation and the [Na+]/[K+] ratio in seedlings challenged by salinity stress. [Na+] in (A) the root and (B) the shoot. The [Na+]/[K+] ratio in (C) the root and (D) the shoot. Values shown in the form mean ± SE (n = 5). Significant differences (P < 0.05) between the test genotype and WT are indicated by an asterisk. ns, non-significant difference. DW, dry weight.
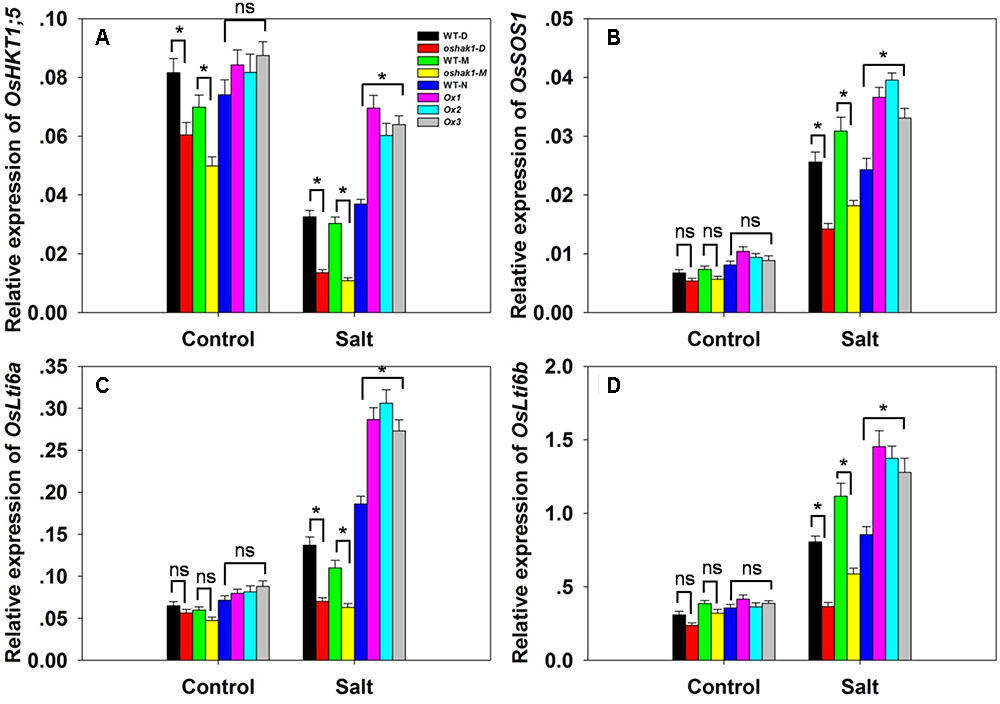
FIGURE 7. The abundance of transcript from genes encoding Na+ transporters. (A–D) The transcription in the roots of hydroponically-grown seedlings raised either in the presence or absence of salinity stress: (A) OsHKT1;5, (B) OsSOS1, (C) OsLti6a, (D) OsLti6b. Outputs derived from qRT-PCRs were normalized against the abundance of Ubq transcript. Values shown in the form mean ± SE (n = 3). Significant differences (P < 0.05) between the test genotype and WT are indicated by an asterisk. ns, non-significant difference.
The Effect of OsHAK1 on K+ and Na+ Homeostasis at the Tillering Stage
When grown in soil, the oshak1 mutants took up substantially more Na+ into both their roots and shoots under control conditions, but the differences in tissue [Na+] and [Na+]/[K+] ratio between the over-expressor and WT plants were not statistically significant (Supplementary Figure 3). In salinized soil, the over-expressor plants accumulated significantly less Na+ in both their shoot and root tissue than did WT plants (Supplementary Figures 3A,B). The [Na+]/[K+] ratio in both the root and shoot was notably lower in the over-expressor plants than in WT ones (Supplementary Figures 3C,D). In contrast, relative to WT plants, the knock-out mutants accumulated about 43% more Na+ in their shoot and about 54% more in their root tissue (Supplementary Figures 3A,B), resulting in a marked increase in the [Na+]/[K+] ratio (Supplementary Figures 3C,D).
The Effect of Varying the Level of OsHAK1 Expression on the Cellular Physiology of Salinity-Stressed Plants
Neither the photosynthetic rate nor the leaf chlorophyll content varied between the genotypes under control conditions (Figures 8A,B). The effect of salinity stress was to reduce the photosynthetic rate in all three genotypes, but the decline was more marked in WT than in OsHAK1 over-expressor plants, resulting in the latter maintaining a significantly higher photosynthetic rate (Figure 8A). Similarly, the leaf chlorophyll content was reduced by the stress in each of the genotypes (Figure 8B). The oshak1 mutants experienced the largest salinity stress-induced fall with respect to both photosynthetic rate and leaf chlorophyll content (Figures 8A,B). The OsHAK1 over-expressor plants accumulated notably less H2O2 in response to salinity stress than did WT ones, while the H2O2 content of the knock-out mutant plants was ∼35% higher than that of their respective WT (Figure 8C). Under control conditions, there was no difference with respect to relative electrolyte leakage between the over-expressor and the WT plants, whereas after exposure to salinity, the over-expressor plants’ leaves suffered a significantly smaller decrease in relative electrolyte leakage than the WT plants’ leaves; the extent of leakage from the mutant plants’ leaves was substantially greater than that from their respective WT (Figure 8D). Similarly, the over-expression of OsHAK1 reduced the accumulation of MDA: under salinity stress, their MDA content was 25% below that of WT plants, while the mutant plants accumulated 45% more than the WT (Figure 8E). When grown under control conditions, the proline content of the tissue of the over-expressor plants was similar to that of WT plants (Figure 8F). The effect of the imposed salinity stress was an increase in proline content in both the over-expressor and WT plants; even so, the over-expressor lines accumulated 35% more proline than the WT plants, whereas the knock-out mutant plants accumulated about 30% less proline than the WT plants (Figure 8F).
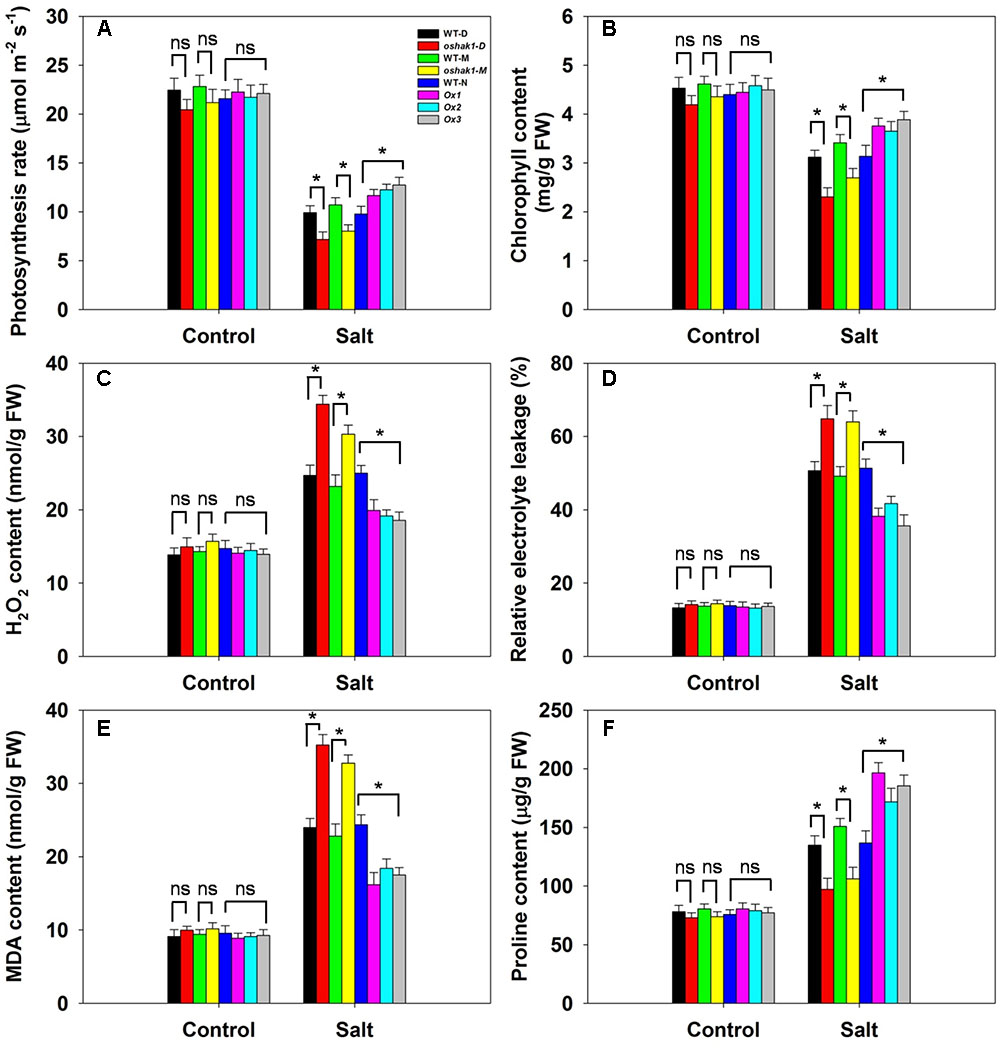
FIGURE 8. The effect of OsHAK1 on the physiology of soil-grown plants exposed to salinity stress. (A) Photosynthetic rate, (B) leaf chlorophyll content, (C) H2O2 content, (D) relative electrolyte leakage, (E) MDA content and (F) proline content. Values shown in the form mean ± SE (n = 5). Significant differences (P < 0.05) between the test genotype and WT are indicated by an asterisk. ns, non-significant; FW, fresh weight.
Salinity Stressed Plants Expressing the OsHAK1 Transgene Display an Altered Transcriptional Profile of Stress-Responsive Genes
The transcription of the stress-responsive genes OsP5CS1, OsDREB2A, OsAP37 and OsERD1 was monitored in WT, OsHAK1 over-expressor and oshak1 mutant plants. Under control conditions, the abundance of transcript did not vary between the WT and the over-expressor plants, but that of OsDREB2A, OsAP37 and OsERD1 was suppressed in the mutant plants (Figures 9A–D). In response to the salinity treatment, each of the four genes was notably up-regulated in the over-expressor plants. In the oshak1 mutant plants, all four genes were less abundantly transcribed than in WT (Figure 9).
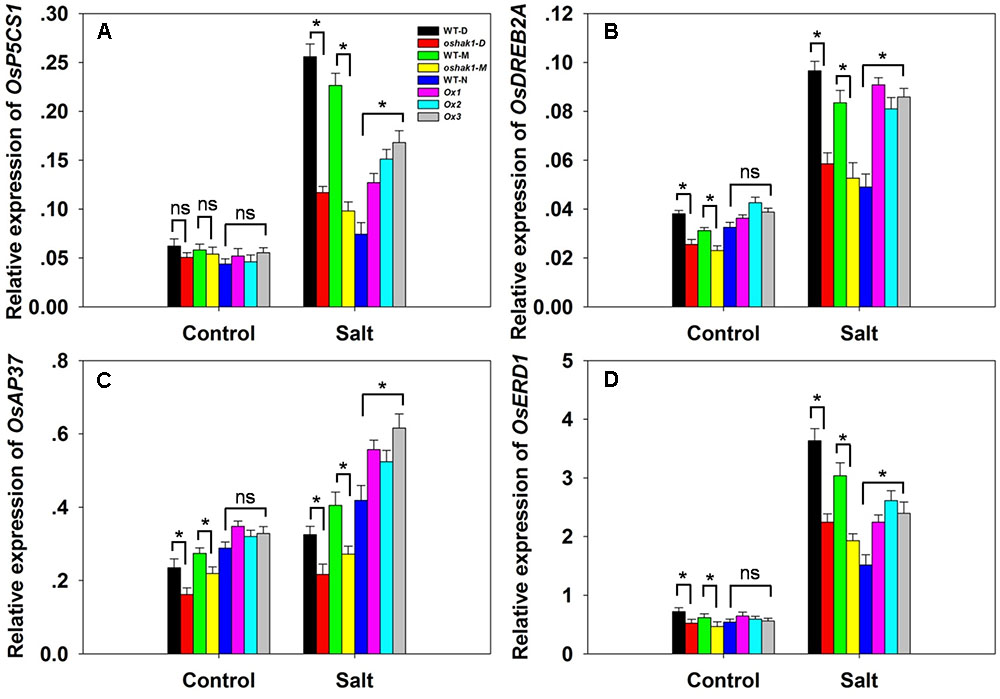
FIGURE 9. The effect of OsHAK1 on the transcription of selected stress-responsive genes. (A–D) The transcription in the youngest two leaves of soil-grown plants in the presence or absence of salinity stress: (A) OsP5CS1, (B) OsDREB2A, (C) OsAP37, (D) OsERD1. Outputs derived from qRT-PCRs were normalized against the abundance of Ubq transcript. Values shown in the form mean ± SE (n = 3). Significant differences (P < 0.05) between the test genotype and WT are indicated by an asterisk. ns, non-significant difference.
The Over-Expression of OsHAK1 Significantly Improved Salinity Tolerance at the Reproductive Stage
OsHAK1 over-expressor and WT plants were subjected to salinity stress at the booting stage through to maturity to assess the tolerance of the OsHAK1 over-expressor when the stress was delayed until the switch from vegetative to reproductive growth. Under control conditions, there was little difference between the agronomic performance of the over-expressor and WT plants (Figures 10A–D). However, when salinity stress was imposed, the over-expressor plants produced 6–15% more effective tillers than the WT plants (Figure 10A), exhibited a 12–18% higher rate of spikelet fertility (Figure 10B), produced grain which were 5–10% heavier (Figure 10C) and yielded 17–35% more per plant (Figure 10D).
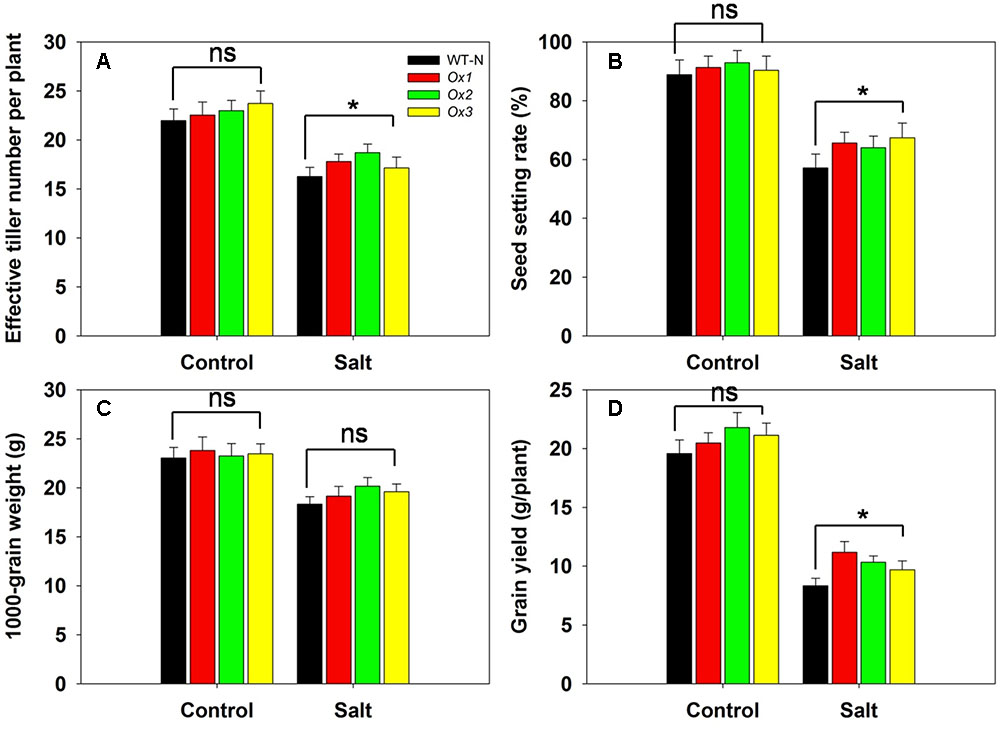
FIGURE 10. The over-expression of OsHAK1 raises the level of rice’s salinity tolerance at the reproductive stage. The stress was applied at the booting stage and maintained through to maturity. (A) Effective tiller number per plant, (B) seed set, (C) 1000 grain weight and (D) grain yield per plant. Values shown in the form mean ± SE (n = 5). Significant differences (P < 0.05) between the over-expressor and WT are indicated by an asterisk. ns, non-significant difference.
Discussion
In rice, OsHAK1 maintains K+-mediated growth and is important for K+ acquisition and transport over a wide range of external [K+] (Chen et al., 2015b). As yet, its participation in the salinity stress response is unclear. A number of lines of evidence generated from the present experiments has revealed that OsHAK1 also makes a positive contribution to salinity tolerance. OsHAK1 was more strongly transcribed in the more tolerant cv. 9311 than in cv. Nipponbare; it was inducible by salinity stress; changes in its transcript abundance correlated positively with tolerance at both the seedling and tillering stages, as well as with K+ and Na+ homeostasis and correlated negatively with lipid peroxidation; its transcription level affected the abundance of several stress-related genes, and finally its over-expression improved the productivity of plants challenged with salinity.
The Up-regulation of OsHAK1 Contributes to the Salinity Tolerance of cv. 9311
Considerable genetic variability for salinity tolerance has been identified in surveys of rice germplasm (Rahman M.A. et al., 2016). Both cv. Nipponbare and the two selected CSSLs proved to be more sensitive to salinity than was cv. 9311: their seedling root and shoot growth was inhibited more strongly, and their leaves suffered a greater degree of chlorosis (Figures 1A,B). A key component of salinity tolerance relates to Na+ and K+ homeostasis (Babu et al., 2017). The [K+] of salinity-stressed cv. 9311 root and shoot tissue was markedly higher than that of both cv. Nipponbare and the two selected CSSLs (Figure 1D), mirroring the observation made by Lee et al. (2003) that tolerant indica varieties feature a greater capacity to take up K+ and to limit the [Na+]/[K+] ratio in the shoot when plants are exposed to salinity stress. An adequate shoot and root cell [K+] is important for the maintenance of growth, as K+ is needed both to maintain the cell turgor needed to ensure cell expansion, and to act as a co-factor for numerous enzymes (Leigh and Wyn Jones, 1984). The two selected CSSLs differ genetically along chromosome 4; the critical difference was related by fine mapping to a 95 Kbp stretch, which harbors the gene OsHAK1. Of all of the 11 genes present in the segment, this was the only one which was differentially transcribed between cvs Nipponbare and 9311, especially when the plants were subjected to salinity stress (Figure 1F). The correlation between K+/Na+ homeostasis and the abundance of OsHAK1 transcript suggests the possibility that OsHAK1 is a key gene in the determination of salinity tolerance, as also proposed by Chen et al. (2015b).
OsHAK1 Regulates K+ and Na+ Homeostasis to Enhance Salinity Stress Resistance
The ability of a plant to restrict the accumulation of Na+ is a central feature of salinity tolerance (Noreen et al., 2010; Cui et al., 2016). The OsHAK1 over-expressors accumulated less Na+ and were thus able to maintain a lower [Na+]/[K+] ratio than could either WT or the oshak1 knock-out mutant (Figures 6A–D and Supplementary Figures 3A–D). The greater accumulation of Na+ in the mutant probably reflected a combination of a higher rate of Na+ uptake by its roots, a higher rate of passive diffusion resulting from compromise to its cell membranes and a less efficient mechanism of sodium efflux (Islam et al., 2015). The over-expression of OsHAK1 may therefore primarily enhance seedling growth under conditions of salinity stress by limiting the uptake of Na+ (Figures 5, 6A–D and Supplementary Figures 3A–D). An ability to control the transport of Na+ from the root to the shoot is also considered to be a significant component of tolerance (Apel and Hirt, 2004; Foyer and Noctor, 2005; Munns and Tester, 2008; Cui et al., 2016). The OsHAK1 over-expressors accumulated less Na+ in their shoot tissue (Figures 5, 6B), thereby limiting the [Na+]/[K+] ratio (Figures 6C,D); this parameter is particularly critical for ensuring normal cell function under conditions of salinity stress (Su et al., 2015). The active removal of Na+ is thought to be controlled by the products of genes belonging to the HKT family (Hamamoto et al., 2015). For example, an RNAi-induced knock-down of the wheat gene TaHKT1;5 results in an increased accumulation of Na+ in the leaves (Byrt et al., 2014). For this reason it was of interest to assay the expression of its rice homolog in the present materials: the indication was that the gene was down-regulated in the oshak1 mutant, which can explain why the mutant accumulated more Na+ in its tissue than did the OsHAK1 over-expressor plants (Figures 5, 6A,B, 7A).
PMP3 (plasma membrane protein 3) is a conserved hydrophobic protein expressed in response to a variety of abiotic stresses (Chidambaram et al., 2015). In yeast, the loss of PMP3 function promotes Na+ accumulation, enhances sensitivity to salinity and induces membrane hyperpolarization (Nylander et al., 2001). In both A. thaliana and Avicennia marina, raising the abundance of this protein has been associated with a reduction in shoot [Na+] and a partial alleviation of stress-induced growth inhibition (Mitsuya et al., 2006; Chidambaram et al., 2015). The rice genome harbors two genes encoding PMP3 homologs, namely OsLti6a and OsLti6b. When plants were exposed to salinity stress, both of these genes were transcribed at a lower level in the oshak1 mutant than in WT (Figures 7C,D). According to Mekawy et al. (2015), PMP3 gene products act to control the uptake of Na+ by the roots of a tolerant cultivar, thereby limiting its leaf [Na+]. The up-regulation of the two OsLti6 genes in the OsHAK1 over-expressors may be due to the induction of other Na+ transporters, since PMP3 proteins contribute indirectly to cation homeostasis by interacting with other ion transporters (Fu et al., 2012). The plasma membrane Na+/H+ exchanger encoded by SOS1 is considered to be the major plant mediator of Na+ efflux (Kronzucker and Britto, 2011): it makes an important contribution to the maintenance of Na+ and K+ homeostasis (Yamaguchi et al., 2013) and is involved in the movement of Na+ through the xylem (Olías et al., 2009). Here, the up-regulation of OsSOS1 in the OsHAK1 over-expressor plants probably enhanced the plants’ ability to prevent the uptake of Na+, thereby lowering the [Na+]/[K+] ratio in their shoot (Figures 6B, 7B).
The Positive Effect of OsHAK1 on Root and Shoot Growth
The faster a shoot is able to grow, the greater the dilution effect on toxic ions such as Na+. Rapid growth in spite of the presence of salinity stress has been suggested to represent a reliable indicator of tolerance (Yeo and Flowers, 1986; Yeo et al., 1990; Rahman M.A. et al., 2016). Here, the over-expression of OsHAK1 had a positive effect on the growth of both the root and the shoot, in contrast to the negative effect of its loss-of-function in the oshak1 mutant (Figure 5). The boost to growth was not limited to seedlings, as soil-grown OsHAK1 over-expressors stressed post booting also recorded an improved performance with respect to grain yield (Figure 10). The benefit of OsHAK1 over-expression may have been due to its depressive effect on Na+ accumulation, in particular thereby supporting the uptake of K+. Alternatively (or additionally) it may have derived from its protection against chlorophyll loss (Figures 8A,B), and thereby its maintenance of a high photosynthetic rate.
The Improved Salinity Tolerance Derived from OsHAK1 Over-Expression Is Reflected in a Positive Effect on a Number of Traits
The over-expression of OsHAK1 had a measurable effect on a number of physiological parameters: it lowered the cellular content of H2O2 (Figure 8C) and MDA (Figure 8E), and promoted that of proline (Figure 8F). A well-documented effect of salinity stress in rice is the accumulation of H2O2 (Mostofa et al., 2015; Rahman A. et al., 2016), so the over-expressor tissue likely experienced a lower level of oxidative damage. Cellular MDA content is commonly used as a surrogate for determining the extent of lipid peroxidation (Zhao et al., 2014); the conclusion that membrane damage was less severe in the OsHAK1 over-expressor than in WT was supported by its reduced extent of electrolyte leakage (Figure 8D). Proline has long been considered to act as an osmoprotectant (Yancey et al., 1982), but its contribution to osmoregulaton in salinity-stressed rice is controversial (Lutts et al., 1996). Here, when exposed to salinity stress, the OsHAK1 over-expressors accumulated significantly more proline than WT (Figure 8F). The product of the gene P5CS1, which is implicated in proline synthesis, has been identified by Zhu et al. (1998) as being a key component of abiotic stress tolerance. The gene can be induced by a variety of stress agents, resulting in an increased proline content, while its over-expression in rice has been shown to improve osmotolerance (Igarashi et al., 1997; Zhu et al., 1998). Here, the higher abundance of OsP5CS1 transcript in tissue of the stressed OsHAK1 over-expressors was fully consistent with the observed increase in their tissue proline content (Figures 8F, 9A). It is possible that the extra proline could also have improved the plant’s capacity to retain water (Song et al., 2012). Proline has also been suggested to act as an antioxidant (Székely et al., 2008), so a higher proline content may have also contributed to limiting electrolyte leakage and lowering the MDA content (Figures 8D–F).
The qRT-PCR analysis revealed a correlation between the transcript abundance of certain stress-related genes and the expression of salinity tolerance in the OsHAK1 over-expressors. The over-expression of a number of transcription factors has been shown to up-regulate various stress-related genes, an effect suggested to contribute toward an improved level of stress tolerance (Kasuga et al., 1999; Mukhopadhyay et al., 2004; Zhao et al., 2014; Hong et al., 2016; Jiang et al., 2016; Shen et al., 2017). Here, the transcription of four such stress-related genes was shown to be enhanced in salinity-challenged OsHAK1 over-expressors (Figure 9). One of these was P5CS1, as mentioned previously. The second was DREB2A, a gene which encodes a transcription factor known to influence both salinity and dehydration stress tolerance (Cui et al., 2011; Mallikarjuna et al., 2011). The third was AP37, encoding a protein associated with tolerance to drought, salinity and low temperature at the vegetative stage and with grain yield under severe drought (Oh et al., 2009). The fourth was a homolog of the A. thaliana gene AtERD1, which is inducible by dehydration stress (Kiyosue et al., 1994); the OsERD1 gene is significantly up-regulated in a rice plant over-expressing a gene encoding a NAC transcription factor (ONAC022), which displays an enhanced level of salinity tolerance (Hong et al., 2016). The suggestion is that the enhanced tolerance to salinity stress generated by OsHAK1 over-expression is, at least in part, a consequence of the up-regulation of these genes. ABA signaling has long been implicated as an important trigger of the plant response to various abiotic agents, as it regulates a number of key stress-related genes (Zhao et al., 2014). The salinity induced up-regulation in the OsHAK1 over-expressors of both early response genes such as DREB2A and AP37, and of late ones such as P5CS1 (Figure 9) implies that the OsHAK1 product participates in ABA signal transduction either directly or indirectly. As yet the mechanistic basis of this involvement is unclear.
Conclusion
The present study has demonstrated that variation in the intensity of OsHAK1 expression can explain a part of the difference in the salinity tolerance expressed by cvs 9311 and Nipponbare. When up-regulated, OsHAK1 improves the tolerance of the rice plant to salinity stress by regulating K+ and Na+ homeostasis, by promoting the growth of the root and shoot, by favoring the accumulation of proline, by protecting against stress-induced damage to the plasma membranes and by activating a number of stress-related genes. The over-expression of many genes in rice, while potentially addressing a trait weakness (such as salinity tolerance), frequently has undesirable pleiotropic effects on development and/or productivity (Kasuga et al., 1999; Hsieh et al., 2002; Nakashima et al., 2007; Morran et al., 2011). The over-expression of OsHAK1 did not, however, appear to induce any such effects on plants grown in the absence of salinity stress (Figures 5, 10). Most promisingly, the over-expressor expressed its enhanced salinity tolerance under field conditions, recording a 25% increase in grain yield per plant compared to the WT (Figure 10D). The implication is that OsHAK1 represents a favorable gene target for the breeding of rice cultivars able to maintain productivity in saline-affected soils. It would be of interest to test the effect of transferring the OsHAK1 allele present in a salinity tolerant cultivar to a salinity sensitive one, in particular to determine whether such an allelic exchange would alter the K+ and Na+ homeostasis in a way which would enhance its salinity tolerance.
Author Contributions
Conceived and designed the experiments: GC, GX, and QQ. Performed the experiments: GC, CL, YZ, and AZ. Analyzed the data: GC, CL, ZG, GX, and QQ. Contributed reagents/materials/analysis tools: LZ, JH, DR, and LY. Wrote and revised the paper: GC, ZG, GX, and QQ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by National Natural Science Foundation of China (No. 31601811) and the Central Level Scientific Research Institutes for the Basic Research and Development Special Fund Business (Grant No. 2015RG001-2).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02216/full#supplementary-material
Footnotes
References
Ahmad, I., Mian, A., and Maathuis, F. J. (2016). Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J. Exp. Bot. 67, 2689–2698. doi: 10.1093/jxb/erw103
Ahmadi, N., Negrão, S., Katsantonis, D., Frouin, J., Ploux, J., Letourmy, P., et al. (2011). Targeted association analysis identified japonica rice varieties achieving Na+/K+ homeostasis without the allelic make-up of the salt tolerant indica variety Nona Bokra. Theoret. Appl. Genet. 123, 881–895. doi: 10.1007/s00122-011-1634-4
Alemán, F., Caballero, F., Ródenas, R., Rivero, R. M., Martínez, V., and Rubio, F. (2014). The F130S point mutation in the Arabidopsis high-affinity K+ transporter AtHAK5 increases K+ over Na+ and Cs+ selectivity and confers Na+ and Cs+ tolerance to yeast under heterologous expression. Front. Plant Sci. 5:430. doi: 10.3389/fpls.2014.00430
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Babu, N. N., Krishnan, S. G., Vinod, K. K., Krishnamurthy, S. L., Singh, V. K., Singh, M. P., et al. (2017). Marker aided incorporation of Saltol, a major QTL associated with seedling stage salt tolerance, into Oryza sativa ‘Pusa Basmati 1121’. Front. Plant Sci. 8:41. doi: 10.3389/fpls.2017.00041
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Byrt, C. S., Xu, B., Krishnan, M., Lightfoot, D. J., Athman, A., Jacobs, A. K., et al. (2014). The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J. 80, 516–526. doi: 10.1111/tpj.12651
Campbell, M. T., Bandillo, N., Al Shiblawi, F. R. A., Sharma, S., Liu, K., Du, Q., et al. (2017). Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLOS Genet. 13:e1006823. doi: 10.1371/journal.pgen.1006823
Chen, G., Feng, H., Hu, Q., Qu, H., Chen, A., Yu, L., et al. (2015a). Improving rice tolerance to potassium deficiency by enhancing OsHAK16p:WOX11-controlled root development. Plant Biotechnol. J. 13, 833–848. doi: 10.1111/pbi.12320
Chen, G., Hu, Q., Luo, L. E., Yang, T., Zhang, S., Hu, Y., et al. (2015b). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38, 2747–2765. doi: 10.1111/pce.12585
Chen, G., Liu, C., Gao, Z., Zhang, Y., Jiang, H., Zhu, L., et al. (2017). OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice. Front. Plant Sci. 8:1885. doi: 10.3389/fpls.2017.01885
Chen, G., Liu, C., Gao, Z., Zhang, Y., Zhu, L., Hu, J., et al. (2018). Driving the expression of RAA1 with a drought-responsive promoter enhances root growth in rice, its accumulation of potassium and its tolerance to moisture stress. Environ. Exp. Bot. 147, 147–156. doi: 10.1016/j.envexpbot.2017.12.008
Chidambaram, R., Venkataraman, G., and Parida, A. (2015). Analysis of transcriptional regulation and tissue-specific expression of Avicennia marina plasma membrane protein 3 suggests it contributes to Na+ transport and homoeostasis in A. marina. Plant Sci. 236, 89–102. doi: 10.1016/j.plantsci.2015.03.013
Cui, M., Zhang, W., Zhang, Q., Xu, Z., Zhu, Z., Duan, F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49, 1384–1391. doi: 10.1016/j.plaphy.2011.09.012
Cui, P., Liu, H., Islam, F., Li, L., Farooq, M. A., Ruan, S., et al. (2016). OsPEX11, a peroxisomal biogenesis factor 11, contributes to salt stress tolerance in Oryza sativa. Front. Plant Sci. 7:1357. doi: 10.3389/fpls.2016.01357
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Fageria, N. K. (1985). Salt tolerance of rice cultivars. Plant Soil 88, 237–243. doi: 10.1007/BF02182450
Flowers, T. J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55, 307–319. doi: 10.1093/jxb/erh003
Flowers, T. J., and Yeo, A. R. (1995). Breeding for salinity resistance in crop plants: where next? Funct. Plant Biol. 22, 875–884. doi: 10.1071/PP9950875
Foyer, C. H., and Noctor, G. (2005). Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 28, 1056–1071. doi: 10.1111/j.1365-3040.2005.01327.x
Fu, J., Zhang, D. F., Liu, Y. H., Ying, S., Shi, Y. S., Song, Y. C., et al. (2012). Isolation and characterization of maize PMP3 genes involved in salt stress tolerance. PLOS ONE 7:e31101. doi: 10.1371/journal.pone.0031101
Gregorio, G. B., Senadhira, D., Mendoza, R. D., Manigbas, N. L., Roxas, J. P., and Guerta, C. Q. (2002). Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 76, 91–101. doi: 10.1016/S0378-4290(02)00031-X
Hamamoto, S., Horie, T., Hauser, F., Deinlein, U., Schroeder, J. I., and Uozumi, N. (2015). HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr. Opin. Biotechnol. 32, 113–120. doi: 10.1016/j.copbio.2014.11.025
Hong, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4. doi: 10.3389/fpls.2016.00004
Horie, T., Brodsky, D. E., Costa, A., Kaneko, T., Schiavo, F. L., Katsuhara, M., et al. (2011a). K+ transport by the OsHKT2; 4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol. 156, 1493–1507. doi: 10.1104/pp.110.168047
Horie, T., Sugawara, M., Okada, T., Taira, K., Kaothien-Nakayama, P., Katsuhara, M., et al. (2011b). Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioengin. 111, 346–356. doi: 10.1016/j.jbiosc.2010.10.014
Hsieh, T. H., Lee, J. T., Charng, Y. Y., and Chan, M. T. (2002). Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 130, 618–626. doi: 10.1104/pp.006783
Igarashi, Y., Yoshiba, Y., Sanada, Y., Yamaguchi-Shinozaki, K., Wada, K., and Shinozaki, K. (1997). Characterization of the gene for Δ1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol. Biol. 33, 857–865. doi: 10.1023/A:1005702408601
Islam, F., Yasmeen, T., Ali, S., Ali, B., Farooq, M. A., and Gill, R. A. (2015). Priming-induced antioxidative responses in two wheat cultivars under saline stress. Acta Physiol. Plant. 37:153. doi: 10.1007/s11738-015-1897-5
Jiang, Y., Qiu, Y., Hu, Y., and Yu, D. (2016). Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front. Plant Sci. 7:145. doi: 10.3389/fpls.2016.00145
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Khan, M. S. A., Hamid, A., and Karim, M. A. (1997). Effect of sodium chloride on germination and seedling characters of different types of rice (Oryza sativa L.). J. Agron. Crop Sci. 179, 163–169. doi: 10.1111/j.1439-037X.1997.tb00512.x
Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol. Biol. 25, 791–798. doi: 10.1007/BF00028874
Koyama, M. L., Levesley, A., Koebner, R. M., Flowers, T. J., and Yeo, A. R. (2001). Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol. 125, 406–422. doi: 10.1104/pp.125.1.406
Kronzucker, H. J., and Britto, D. T. (2011). Sodium transport in plants: a critical review. New Phytol. 189, 54–81. doi: 10.1111/j.1469-8137.2010.03540.x
Lee, K. S., Choi, W. Y., Ko, J. C., Kim, T. S., and Gregorio, G. B. (2003). Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 216, 1043–1046. doi: 10.1007/s00425-002-0958-3
Leigh, R. A., and Wyn Jones, R. G. (1984). A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytologist 97, 1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x
Li, B. Z., Xin, W. J., Sun, S. B., Shen, Q. R., and Xu, G. H. (2006). Physiological and molecular responses of nitrogen-starved rice plants to resupply of different nitrogen sources. Plant Soil 287, 145–159. doi: 10.1007/s11104-006-9051-1
Li, Q., Tang, Z., Hu, Y., Yu, L., Liu, Z., and Xu, G. (2014). Functional analyses of a putative plasma membrane Na+/H+ antiporter gene isolated from salt tolerant Helianthus tuberosus. Mol. Biol. Rep. 41, 5097–5108. doi: 10.1007/s11033-014-3375-3
Li, Q., Yang, A., and Zhang, W. H. (2016). Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L.). J. Exp. Bot. 67, 6431–6444. doi: 10.1093/jxb/erw407
Li, Y., Gu, M., Zhang, X., Zhang, J., Fan, H. M., Li, P. P., et al. (2014). Engineering a sensitive visual tracking reporter system for real-time monitoring phosphorus deficiency in tobacco. Plant Biotechnol. J. 12, 674–684. doi: 10.1111/pbi.12171
Lin, H. X., Zhu, M. Z., Yano, M., Gao, J. P., Liang, Z. W., Su, W. A., et al. (2004). QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 108, 253–260. doi: 10.1007/s00122-003-1421-y
Luan, S., Lan, W., and Chul Lee, S. (2009). Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL–CIPK network. Curr. Opin. Plant Biol. 12, 339–346. doi: 10.1016/j.pbi.2009.05.003
Lutts, S., Kinet, J. M., and Bouharmont, J. (1995). Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 46, 1843–1852. doi: 10.1093/jxb/46.12.1843
Lutts, S., Kinet, J. M., and Bouharmont, J. (1996). Effects of salt stress on growth, mineral nutrition and proline accumulation in relation to osmotic adjustment in rice (Oryza sativa L.) cultivars differing in salinity resistance. Plant Growth Regulat. 19, 207–218. doi: 10.1007/BF00037793
Maathuis, F. J. (2006). The role of monovalent cation transporters in plant responses to salinity. J. Exp. Bot. 57, 1137–1147. doi: 10.1093/jxb/erj001
Mallikarjuna, G., Mallikarjuna, K., Reddy, M. K., and Kaul, T. (2011). Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol. Lett. 33, 1689–1697. doi: 10.1007/s10529-011-0620-x
Martínez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J. K., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012. doi: 10.1104/pp.106.092635
Mekawy, A. M. M., Assaha, D. V., Yahagi, H., Tada, Y., Ueda, A., and Saneoka, H. (2015). Growth, physiological adaptation, and gene expression analysis of two Egyptian rice cultivars under salt stress. Plant Physiol. Biochem. 87, 17–25. doi: 10.1016/j.plaphy.2014.12.007
Mitsuya, S., Taniguchi, M., Miyake, H., and Takabe, T. (2006). Overexpression of RCI2A decreases Na+ uptake and mitigates salinity-induced damages in Arabidopsis thaliana plants. Physiol. Plant. 128, 95–102. doi: 10.1111/j.1399-3054.2006.00714.x
Morran, S., Eini, O., Pyvovarenko, T., Parent, B., Singh, R., Ismagul, A., et al. (2011). Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. doi: 10.1111/j.1467-7652.2010.00547.x
Mostofa, M. G., and Fujita, M. (2013). Salicylic acid alleviates copper toxicity in rice (Oryza sativa L.) seedlings by up-regulating antioxidative and glyoxalase systems. Ecotoxicology 22, 959–973. doi: 10.1007/s10646-013-1073-x
Mostofa, M. G., Saegusa, D., Fujita, M., and Tran, L. S. P. (2015). Hydrogen sulfide regulates salt tolerance in rice by maintaining Na+/K+ balance, mineral homeostasis and oxidative metabolism under excessive salt stress. Front. Plant Sci. 6:1055. doi: 10.3389/fpls.2015.01055
Mukhopadhyay, A., Vij, S., and Tyagi, A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 101, 6309–6314. doi: 10.1073/pnas.0401572101
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nakashima, K., Tran, L. S. P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Negrão, S., Courtois, B., Ahmadi, N., Abreu, I., Saibo, N., and Oliveira, M. M. (2011). Recent updates on salinity stress in rice: from physiological to molecular responses. Crit. Rev. Plant Sci. 30, 329–377. doi: 10.1080/07352689.2011.587725
Noreen, Z., Ashraf, M., and Akram, N. A. (2010). Salt-induced regulation of some key antioxidant enzymes and physio-biochemical phenomena in five diverse cultivars of turnip (Brassica rapa L.). J. Agron. Crop Sci. 196, 273–285.
Nylander, M., Heino, P., Helenius, E., Palva, E. T., Ronne, H., and Welin, B. V. (2001). The low-temperature- and salt-induced RCI2A gene of Arabidopsis complements the sodium sensitivity caused by a deletion of the homologous yeast gene SNA1. Plant Mol. Biol. 45, 341–352. doi: 10.1023/A:1006451914231
Oh, S. J., Kim, Y. S., Kwon, C. W., Park, H. K., Jeong, J. S., and Kim, J. K. (2009). Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 150, 1368–1379. doi: 10.1104/pp.109.137554
Olías, R., Eljakaoui, Z., Li, J., De Morales, P. A., Marín-Manzano, M. C., Pardo, J. M., et al. (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 32, 904–916. doi: 10.1111/j.1365-3040.2009.01971.x
Platten, J. D., Egdane, J. A., and Ismail, A. M. (2013). Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism? BMC Plant Biol. 13:32. doi: 10.1186/1471-2229-13-32
Rahman, A., Nahar, K., Hasanuzzaman, M., and Fujita, M. (2016). Calcium supplementation improves Na+/K+ ratio, antioxidant defense and glyoxalase systems in salt-stressed rice seedlings. Front. Plant Sci. 7:609. doi: 10.3389/fpls.2016.00609
Rahman, M. A., Thomson, M. J., Shah-E-Alam, M., de Ocampo, M., Egdane, J., and Ismail, A. M. (2016). Exploring novel genetic sources of salinity tolerance in rice through molecular and physiological characterization. Ann. Bot. 117, 1083–1097. doi: 10.1093/aob/mcw030
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Santa-María, G. E., Rubio, F., Dubcovsky, J., and Rodríguez-Navarro, A. (1997). The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9, 2281–2289. doi: 10.1105/tpc.9.12.2281
Schachtman, D., and Liu, W. (1999). Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 4, 281–287. doi: 10.1016/S1360-1385(99)01428-4
Shen, J., Lv, B., Luo, L., He, J., Mao, C., Xi, D., et al. (2017). The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 7:40641. doi: 10.1038/srep40641
Song, S., Chen, Y., Zhao, M., and Zhang, W. H. (2012). A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses. Environ. Exp. Bot. 80, 1–9. doi: 10.1016/j.envexpbot.2012.02.001
Sripinyowanich, S., Klomsakul, P., Boonburapong, B., Bangyeekhun, T., Asami, T., Gu, H., et al. (2013). Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ. Exp. Bot. 86, 94–105. doi: 10.1016/j.envexpbot.2010.01.009
Su, Y., Luo, W., Lin, W., Ma, L., and Kabir, M. H. (2015). Model of cation transportation mediated by high-affinity potassium transporters (HKTs) in higher plants. Biol. Proced. Online 17, 484–488.
Székely, G., Ábrahám, E., Cséplö,Á., Rigó, G., Zsigmond, L., Csiszár, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Takahashi, R., Nishio, T., Ichizen, N., and Takano, T. (2007). High-affinity K+ transporter PhaHAK5 is expressed only in salt-sensitive reed plants and shows Na+ permeability under NaCl stress. Plant Cell Rep. 26, 1673–1679. doi: 10.1007/s00299-007-0364-1
Thomson, M. J., Ocampo, M., Egdane, J., Rahman, M. A., Sajise, A. G., Adorada, D. L., et al. (2010). Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3, 148–160. doi: 10.1007/s12284-010-9053-8
Xu, H., Jiang, X., Zhan, K., Cheng, X., Chen, X., Pardo, J. M., et al. (2008). Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 473, 8–15. doi: 10.1016/j.abb.2008.02.018
Yamaguchi, T., Hamamoto, S., and Uozumi, N. (2013). Sodium transport system in plant cells. Front. Plant Sci. 4:410. doi: 10.3389/fpls.2013.00410
Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D., and Somero, G. N. (1982). Living with water stress: evolution of osmolyte systems. Science 217, 1214–1222. doi: 10.1126/science.7112124
Yang, T., Zhang, S., Hu, Y., Wu, F., Hu, Q., Chen, G., et al. (2014). The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166, 945–959. doi: 10.1104/pp.114.246520
Yeo, A. R., and Flowers, T. J. (1986). Salinity resistance in rice (Oryza sativa L.) and a pyramiding approach to breeding varieties for saline soils. Funct. Plant Biol. 13, 161–173.
Yeo, A. R., Yeo, M. E., Flowers, S. A., and Flowers, T. J. (1990). Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor. Appl. Genet. 79, 377–384. doi: 10.1007/BF01186082
Zeng, L., Shannon, M. C., and Grieve, C. M. (2002). Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica 127, 235–245. doi: 10.1023/A:1020262932277
Zeng, Y., Li, Q., Wang, H., Zhang, J., Du, J., Feng, H., et al. (2017). Two NHX-type transporters from Helianthus tuberosus improve the tolerance of rice to salinity and nutrient deficiency stress. Plant Biotechnol. J. doi: 10.1111/pbi.12773 [Epub ahead of print].
Zhao, Y., Ma, Q., Jin, X., Peng, X., Liu, J., Deng, L., et al. (2014). A novel maize homeodomain–leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 55, 1142–1156. doi: 10.1093/pcp/pcu054
Keywords: salinity stress, OsHAK1, Na+ and K+ homeostasis, Oryza sativa, 9311, Nipponbare
Citation: Chen G, Liu C, Gao Z, Zhang Y, Zhang A, Zhu L, Hu J, Ren D, Yu L, Xu G and Qian Q (2018) Variation in the Abundance of OsHAK1 Transcript Underlies the Differential Salinity Tolerance of an indica and a japonica Rice Cultivar. Front. Plant Sci. 8:2216. doi: 10.3389/fpls.2017.02216
Received: 07 September 2017; Accepted: 18 December 2017;
Published: 05 January 2018.
Edited by:
Chengdao Li, Murdoch University, AustraliaCopyright © 2018 Chen, Liu, Gao, Zhang, Zhang, Zhu, Hu, Ren, Yu, Xu and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Qian, qianqian188@hotmail.com Guohua Xu, ghxu@njau.edu.cn Guang Chen, chenguang0066@126.com
 Guang Chen
Guang Chen Chaolei Liu
Chaolei Liu Zhenyu Gao
Zhenyu Gao Yu Zhang
Yu Zhang Anpeng Zhang1
Anpeng Zhang1 Deyong Ren
Deyong Ren Guohua Xu
Guohua Xu Qian Qian
Qian Qian