Herpesviral Latency—Common Themes
Abstract
:1. Introduction
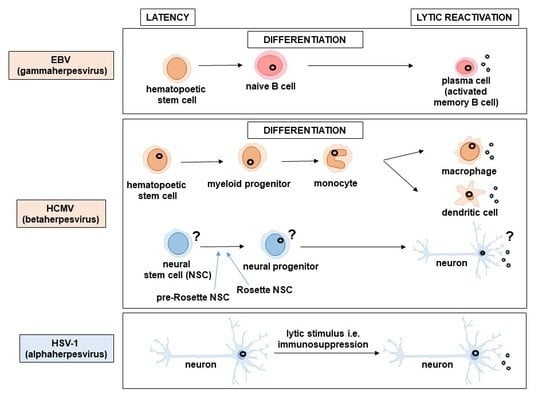
2. Latent Genome Persistence
3. Viral Gene Expression Patterns in Latency
4. Virus Reactivation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pellett, P.E.; Roizman, B. Herpesviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Speck, S.H.; Ganem, D. Viral latency and its regulation: Lessons from the gamma-herpesviruses. Cell Host Microbe 2010, 8, 100–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roizman, B.; Knipe, D.M.; Whitley, R.J. Herpes Simplex Viruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Spear, P.G.; Longnecker, R. Herpesvirus entry: An update. J. Virol. 2003, 77, 10179–10185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitley, R.J. Herpesviruses. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Mocarski, E.S.; Shenk, T.; Pass, R.F. Cytomegalovirus. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Davison, A.J. Herpesviruses: General Features. Ref. Modul. Biomed. Sci. 2014. [Google Scholar] [CrossRef]
- Hahn, G.; Jores, R.; Mocarski, E.S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 1998, 95, 3937–3942. [Google Scholar] [CrossRef] [Green Version]
- Luppi, M.; Barozzi, P.; Morris, C.; Maiorana, A.; Garber, R.; Bonacorsi, G.; Donelli, A.; Marasca, R.; Tabilio, A.; Torelli, G. Human herpesvirus 6 latently infects early bone marrow progenitors in vivo. J. Virol. 1999, 73, 754–759. [Google Scholar] [CrossRef] [Green Version]
- Belzile, J.P.; Stark, T.J.; Yeo, G.W.; Spector, D.H. Human cytomegalovirus infection of human embryonic stem cell-derived primitive neural stem cells is restricted at several steps but leads to the persistence of viral DNA. J. Virol. 2014, 88, 4021–4039. [Google Scholar] [CrossRef] [Green Version]
- Means, R.E.; Lang, S.M.; Jung, J.U. Human gammaherpesvirus immune evasion strategies. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, 1st ed.; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Parravicini, C.; Chandran, B.; Corbellino, M.; Berti, E.; Paulli, M.; Moore, P.S.; Chang, Y. Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am. J. Pathol. 2000, 156, 743–749. [Google Scholar] [CrossRef]
- Jha, H.C.; Banerjee, S.; Robertson, E.S. The Role of Gammaherpesviruses in Cancer Pathogenesis. Pathogens 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Jacob, R.J.; Morse, L.S.; Roizman, B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J. Virol. 1979, 29, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Poffenberger, K.L.; Roizman, B. A noninverting genome of a viable herpes simplex virus 1: Presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 1985, 53, 587–595. [Google Scholar] [CrossRef] [Green Version]
- Luppi, M.; Marasca, R.; Barozzi, P.; Ferrari, S.; Ceccherini-Nelli, L.; Batoni, G.; Merelli, E.; Torelli, G. Three cases of human herpesvirus-6 latent infection: Integration of viral genome in peripheral blood mononuclear cell DNA. J. Med. Virol. 1993, 40, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Daibata, M.; Taguchi, T.; Taguchi, H.; Miyoshi, I. Integration of human herpesvirus 6 in a Burkitt’s lymphoma cell line. Br. J. Haematol. 1998, 102, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Nacheva, E.P.; Ward, K.N.; Brazma, D.; Virgili, A.; Howard, J.; Leong, H.N.; Clark, D.A. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J. Med. Virol. 2008, 80, 1952–1958. [Google Scholar] [CrossRef] [PubMed]
- Ward, K.N.; Leong, H.N.; Nacheva, E.P.; Howard, J.; Atkinson, C.E.; Davies, N.W.; Griffiths, P.D.; Clark, D.A. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral DNA in blood, sera, and hair follicles. J. Clin. Microbiol. 2006, 44, 1571–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka-Taya, K.; Sashihara, J.; Kurahashi, H.; Amo, K.; Miyagawa, H.; Kondo, K.; Okada, S.; Yamanishi, K. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J. Med. Virol. 2004, 73, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, J.H.; Medveczky, M.M.; Luka, J.; Hadley, S.H.; Luegmayr, A.; Ablashi, D.; Lund, T.C.; Tolar, J.; De Meirleir, K.; Montoya, J.G.; et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 5563–5568. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.C.; Choudhuri, T.; Robertson, E.S. The minimal replicator element of the Kaposi’s sarcoma-associated herpesvirus terminal repeat supports replication in a semiconservative and cell-cycle-dependent manner. J. Virol. 2007, 81, 3402–3413. [Google Scholar] [CrossRef] [Green Version]
- Grundhoff, A.; Ganem, D. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 2003, 77, 2779–2783. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Garber, A.C.; Renne, R. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 2002, 76, 11677–11687. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.; Sohn, H.; Lee, D.; Gwack, Y.; Choe, J. Functional dissection of latency-associated nuclear antigen 1 of Kaposi’s sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 2002, 76, 10320–10331. [Google Scholar] [CrossRef] [Green Version]
- Longnecker, R.; Kieff, E.; Cohen, J.I. Epstein–Barr Virus. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Damania, B.; Cesarman, E. Kaposi’s Sarcoma–Associated Herpesvirus. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2. [Google Scholar]
- Tarrant-Elorza, M.; Rossetto, C.C.; Pari, G.S. Maintenance and replication of the human cytomegalovirus genome during latency. Cell Host Microbe 2014, 16, 43–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, W.; Roizman, B. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 1971, 68, 2818–2821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, J.; Fraser, N.W. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 2008, 82, 3530–3537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groves, I.J.; Reeves, M.B.; Sinclair, J.H. Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic ‘pre-immediate-early’ repression of viral gene expression mediated by histone post-translational modification. J. Gen. Virol. 2009, 90, 2364–2374. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.E.; Levinger, L.F.; Carter, C.W. Nucleosomal structure of Epstein–Barr virus DNA in transformed cell lines. J. Virol. 1979, 29, 657–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wensing, B.; Stühler, A.; Jenkins, P.; Hollyoake, M.; Karstegl, C.E.; Farrell, P.J. Variant chromatin structure of the oriP region of Epstein–Barr virus and regulation of EBER1 expression by upstream sequences and oriP. J. Virol. 2001, 75, 6235–6241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyson, P.J.; Farrell, P.J. Chromatin structure of Epstein–Barr virus. J. Gen. Virol. 1985, 66 Pt 9, 1931–1940. [Google Scholar] [CrossRef]
- Deshmane, S.L.; Fraser, N.W. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 1989, 63, 943–947. [Google Scholar] [CrossRef] [Green Version]
- Stedman, W.; Deng, Z.; Lu, F.; Lieberman, P.M. ORC, MCM, and histone hyperacetylation at the Kaposi’s sarcoma-associated herpesvirus latent replication origin. J. Virol. 2004, 78, 12566–12575. [Google Scholar] [CrossRef] [Green Version]
- Thorley-Lawson, D.A.; Duca, K.A.; Shapiro, M. Epstein–Barr virus: A paradigm for persistent infection—For real and in virtual reality. Trends Immunol. 2008, 29, 195–201. [Google Scholar] [CrossRef]
- Speck, S.H. Regulation of EBV Latency-Associated Gene Expression. In Epstein–Barr Virus; Robertson, E.S., Ed.; Caister Academic Press: Wymondham, UK, 2005. [Google Scholar]
- Yates, J.L.; Warren, N.; Sugden, B. Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature 1985, 313, 812–815. [Google Scholar] [CrossRef] [PubMed]
- Reisman, D.; Yates, J.; Sugden, B. A putative origin of replication of plasmids derived from Epstein–Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 1985, 5, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Sugden, B.; Warren, N. A promoter of Epstein–Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 1989, 63, 2644–2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.A.; Diamond, M.E.; Yates, J.L. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein–Barr virus. J. Virol. 1999, 73, 2974–2982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumi, K.M.; Kieff, E.D. The Epstein–Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl. Acad. Sci. USA 1997, 94, 12592–12597. [Google Scholar] [CrossRef] [Green Version]
- Bornkamm, G.W.; Hammerschmidt, W. Molecular virology of Epstein–Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 437–459. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.L.; Burkhardt, A.L.; Lee, J.H.; Stealey, B.; Longnecker, R.; Bolen, J.B.; Kieff, E. Integral membrane protein 2 of Epstein–Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 1995, 2, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Lerner, M.R.; Andrews, N.C.; Miller, G.; Steitz, J.A. Two small RNAs encoded by Epstein–Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 1981, 78, 805–809. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, S.; Zavolan, M.; Grässer, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef]
- Edwards, R.H.; Marquitz, A.R.; Raab-Traub, N. Epstein–Barr virus BART microRNAs are produced from a large intron prior to splicing. J. Virol. 2008, 82, 9094–9106. [Google Scholar] [CrossRef] [Green Version]
- Toptan, T.; Abere, B.; Nalesnik, M.A.; Swerdlow, S.H.; Ranganathan, S.; Lee, N.; Shair, K.H.; Moore, P.S.; Chang, Y. Circular DNA tumor viruses make circular RNAs. Proc. Natl. Acad. Sci. USA 2018, 115, E8737–E8745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komano, J.; Maruo, S.; Kurozumi, K.; Oda, T.; Takada, K. Oncogenic role of Epstein–Barr virus-encoded RNAs in Burkitt’s lymphoma cell line Akata. J. Virol. 1999, 73, 9827–9831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanbo, A.; Inoue, K.; Adachi-Takasawa, K.; Takada, K. Epstein–Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt’s lymphoma. EMBO J. 2002, 21, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, D.; Zhou, L.; Samanta, M.; Matsumoto, M.; Ebihara, T.; Seya, T.; Imai, S.; Fujieda, M.; Kawa, K.; Takada, K. Epstein–Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J. Exp. Med. 2009, 206, 2091–2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samanta, M.; Iwakiri, D.; Takada, K. Epstein–Barr virus-encoded small RNA induces IL-10 through RIG-I-mediated IRF-3 signaling. Oncogene 2008, 27, 4150–4160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seto, E.; Moosmann, A.; Grömminger, S.; Walz, N.; Grundhoff, A.; Hammerschmidt, W. Micro RNAs of Epstein–Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010, 6, e1001063. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Tsai, M.-H.; Shumilov, A.; Poirey, R.; Bannert, H.; Middeldorp, J.M.; Feederle, R.; Delecluse, H.-J. The Epstein–Barr Virus BART miRNA Cluster of the M81 Strain Modulates Multiple Functions in Primary B Cells. PLoS Pathog. 2015, 11, e1005344. [Google Scholar] [CrossRef]
- Barbera, A.J.; Chodaparambil, J.V.; Kelley-Clarke, B.; Luger, K.; Kaye, K.M. Kaposi’s sarcoma-associated herpesvirus LANA hitches a ride on the chromosome. Cell Cycle 2006, 5, 1048–1052. [Google Scholar] [CrossRef]
- Cotter, M.A.; Robertson, E.S. The latency-associated nuclear antigen tethers the Kaposi’s sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 1999, 264, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Friborg, J.; Kong, W.; Hottiger, M.O.; Nabel, G.J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 1999, 402, 889–894. [Google Scholar] [CrossRef]
- Radkov, S.A.; Kellam, P.; Boshoff, C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 2000, 6, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Fujimuro, M.; Wu, F.Y.; ApRhys, C.; Kajumbula, H.; Young, D.B.; Hayward, G.S.; Hayward, S.D. A novel viral mechanism for dysregulation of beta-catenin in Kaposi’s sarcoma-associated herpesvirus latency. Nat. Med. 2003, 9, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Moore, P.S.; Talbot, S.J.; Boshoff, C.H.; Zarkowska, T.; Godden-Kent; Paterson, H.; Weiss, R.A.; Mittnacht, S. Cyclin encoded by KS herpesvirus. Nature 1996, 382, 410. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.; Ramos da Silva, S.; Bedolla, R.; Ye, F.; Zhou, F.; Gao, S.J. Viral cyclin promotes KSHV-induced cellular transformation and tumorigenesis by overriding contact inhibition. Cell Cycle 2014, 13, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Li, Q.; Lee, J.Y.; Lee, S.H.; Jeong, J.H.; Lee, H.R.; Chang, H.; Zhou, F.C.; Gao, S.J.; Liang, C.; et al. FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 2009, 11, 1355–1362. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, P.M.; Jasmin, A.; Eby, M.T.; Hood, L. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 1999, 18, 5738–5746. [Google Scholar] [CrossRef] [Green Version]
- Sadler, R.; Wu, L.; Forghani, B.; Renne, R.; Zhong, W.; Herndier, B.; Ganem, D. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 1999, 73, 5722–5730. [Google Scholar] [CrossRef] [Green Version]
- Muralidhar, S.; Pumfery, A.M.; Hassani, M.; Sadaie, M.R.; Kishishita, M.; Brady, J.N.; Doniger, J.; Medveczky, P.; Rosenthal, L.J. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) transforming gene. J. Virol. 1998, 72, 4980–4988. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.W.; Tu, P.F.; Hsiao, N.W.; Chang, C.Y.; Wan, L.; Lin, Y.T.; Chang, H.W. Identification of a novel septin 4 protein binding to human herpesvirus 8 kaposin A protein using a phage display cDNA library. J. Virol. Methods 2007, 143, 65–72. [Google Scholar] [CrossRef]
- McCormick, C.; Ganem, D. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science 2005, 307, 739–741. [Google Scholar] [CrossRef]
- Rivas, C.; Thlick, A.E.; Parravicini, C.; Moore, P.S.; Chang, Y. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 2001, 75, 429–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteban, M.; García, M.A.; Domingo-Gil, E.; Arroyo, J.; Nombela, C.; Rivas, C. The latency protein LANA2 from Kaposi’s sarcoma-associated herpesvirus inhibits apoptosis induced by dsRNA-activated protein kinase but not RNase L activation. J. Gen. Virol. 2003, 84, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Joo, C.H.; Shin, Y.C.; Gack, M.; Wu, L.; Levy, D.; Jung, J.U. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi’s sarcoma-associated herpesvirus viral IRF homolog vIRF3. J. Virol. 2007, 81, 8282–8292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandriani, S.; Ganem, D. Array-based transcript profiling and limiting-dilution reverse transcription-PCR analysis identify additional latent genes in Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2010, 84, 5565–5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, P.S.; Boshoff, C.; Weiss, R.A.; Chang, Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 1996, 274, 1739–1744. [Google Scholar] [CrossRef]
- Aoki, Y.; Yarchoan, R.; Wyvill, K.; Okamoto, S.; Little, R.F.; Tosato, G. Detection of viral interleukin-6 in Kaposi sarcoma-associated herpesvirus-linked disorders. Blood 2001, 97, 2173–2176. [Google Scholar] [CrossRef]
- Neipel, F.; Albrecht, J.C.; Fleckenstein, B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: Determinants of its pathogenicity? J. Virol. 1997, 71, 4187–4192. [Google Scholar] [CrossRef] [Green Version]
- Nicholas, J.; Ruvolo, V.R.; Burns, W.H.; Sandford, G.; Wan, X.; Ciufo, D.; Hendrickson, S.B.; Guo, H.G.; Hayward, G.S.; Reitz, M.S. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 1997, 3, 287–292. [Google Scholar] [CrossRef]
- Burger, R.; Neipel, F.; Fleckenstein, B.; Savino, R.; Ciliberto, G.; Kalden, J.R.; Gramatzki, M. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood 1998, 91, 1858–1863. [Google Scholar] [CrossRef]
- Chen, D.; Sandford, G.; Nicholas, J. Intracellular signaling mechanisms and activities of human herpesvirus 8 interleukin-6. J. Virol. 2009, 83, 722–733. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Zhao, Y.; Karijolich, J. The landscape of transcription initiation across latent and lytic KSHV genomes. PLoS Pathog. 2019, 15, e1007852. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.S.; Lee, S.H.; Feng, P.; Chang, H.; Cho, N.H.; Jung, J.U. Characterization of the Kaposi’s sarcoma-associated herpesvirus K1 signalosome. J. Virol. 2005, 79, 12173–12184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Veazey, R.; Williams, K.; Li, M.; Guo, J.; Neipel, F.; Fleckenstein, B.; Lackner, A.; Desrosiers, R.C.; Jung, J.U. Deregulation of cell growth by the K1 gene of Kaposi’s sarcoma-associated herpesvirus. Nat. Med. 1998, 4, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Tang, Z.Y.; Peng, X.; Coleman, R.; Gill, J.; Farr, G.; Samaniego, F. Tumorigenesis and aberrant signaling in transgenic mice expressing the human herpesvirus-8 K1 gene. J. Natl. Cancer Inst. 2002, 94, 926–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomlinson, C.C.; Damania, B. The K1 protein of Kaposi’s sarcoma-associated herpesvirus activates the Akt signaling pathway. J. Virol. 2004, 78, 1918–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Dittmer, D.P.; Tomlinson, C.C.; Fakhari, F.D.; Damania, B. Immortalization of primary endothelial cells by the K1 protein of Kaposi’s sarcoma-associated herpesvirus. Cancer Res 2006, 66, 3658–3666. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Lu, S.; Zhang, Z.; Gonzalez, C.M.; Damania, B.; Cullen, B.R. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 2005, 102, 5570–5575. [Google Scholar] [CrossRef] [Green Version]
- Samols, M.A.; Hu, J.; Skalsky, R.L.; Renne, R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2005, 79, 9301–9305. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.; Lin, S.F.; Gradoville, L.; Miller, G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 1996, 93, 11883–11888. [Google Scholar] [CrossRef] [Green Version]
- Samols, M.A.; Skalsky, R.L.; Maldonado, A.M.; Riva, A.; Lopez, M.C.; Baker, H.V.; Renne, R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007, 3, e65. [Google Scholar] [CrossRef]
- Gottwein, E.; Mukherjee, N.; Sachse, C.; Frenzel, C.; Majoros, W.H.; Chi, J.T.; Braich, R.; Manoharan, M.; Soutschek, J.; Ohler, U.; et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 2007, 450, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Hu, J.; Renne, R. Analysis of viral cis elements conferring Kaposi’s sarcoma-associated herpesvirus episome partitioning and maintenance. J. Virol. 2007, 81, 9825–9837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellare, P.; Ganem, D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: An evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 2009, 6, 570–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, X.; Bai, Z.; Ye, F.; Xie, J.; Kim, C.G.; Huang, Y.; Gao, S.J. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell Biol. 2010, 12, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Goodrum, F.D.; Jordan, C.T.; High, K.; Shenk, T. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: A model for latency. Proc. Natl. Acad. Sci. USA 2002, 99, 16255–16260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, V.T.; Trilling, M.; Hengel, H. The cytomegaloviral protein pUL138 acts as potentiator of tumor necrosis factor (TNF) receptor 1 surface density to enhance ULb’-encoded modulation of TNF-α signaling. J. Virol. 2011, 85, 13260–13270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weekes, M.P.; Tan, S.Y.; Poole, E.; Talbot, S.; Antrobus, R.; Smith, D.L.; Montag, C.; Gygi, S.P.; Sinclair, J.H.; Lehner, P.J. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 2013, 340, 199–202. [Google Scholar] [CrossRef] [Green Version]
- Buehler, J.; Zeltzer, S.; Reitsma, J.; Petrucelli, A.; Umashankar, M.; Rak, M.; Zagallo, P.; Schroeder, J.; Terhune, S.; Goodrum, F. Opposing Regulation of the EGF Receptor: A Molecular Switch Controlling Cytomegalovirus Latency and Replication. PLoS Pathog. 2016, 12, e1005655. [Google Scholar] [CrossRef] [Green Version]
- Bego, M.; Maciejewski, J.; Khaiboullina, S.; Pari, G.; St Jeor, S. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J. Virol. 2005, 79, 11022–11034. [Google Scholar] [CrossRef] [Green Version]
- Keyes, L.R.; Hargett, D.; Soland, M.; Bego, M.G.; Rossetto, C.C.; Almeida-Porada, G.; St Jeor, S. HCMV protein LUNA is required for viral reactivation from latently infected primary CD14⁺ cells. PLoS ONE 2012, 7, e52827. [Google Scholar] [CrossRef]
- Poole, E.L.; Kew, V.G.; Lau, J.C.H.; Murray, M.J.; Stamminger, T.; Sinclair, J.H.; Reeves, M.B. A Virally Encoded DeSUMOylase Activity Is Required for Cytomegalovirus Reactivation from Latency. Cell Rep. 2018, 24, 594–606. [Google Scholar] [CrossRef] [Green Version]
- Beisser, P.S.; Laurent, L.; Virelizier, J.L.; Michelson, S. Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J. Virol. 2001, 75, 5949–5957. [Google Scholar] [CrossRef] [Green Version]
- Cheung, A.K.; Abendroth, A.; Cunningham, A.L.; Slobedman, B. Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood 2006, 108, 3691–3699. [Google Scholar] [CrossRef]
- Humby, M.S.; O’Connor, C.M. Human Cytomegalovirus US28 Is Important for Latent Infection of Hematopoietic Progenitor Cells. J. Virol. 2015, 90, 2959–2970. [Google Scholar] [CrossRef] [Green Version]
- Krishna, B.A.; Poole, E.L.; Jackson, S.E.; Smit, M.J.; Wills, M.R.; Sinclair, J.H. Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection. MBio 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Krishna, B.A.; Humby, M.S.; Miller, W.E.; O’Connor, C.M. Human cytomegalovirus G protein-coupled receptor US28 promotes latency by attenuating c-fos. Proc. Natl. Acad. Sci. USA 2019, 116, 1755–1764. [Google Scholar] [CrossRef] [Green Version]
- Krishna, B.A.; Wass, A.B.; Sridharan, R.; O’Connor, C.M. The Requirement for US28 during Cytomegalovirus Latency Is Independent of US27 and US29 Gene Expression. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, C.; Abendroth, A.; Slobedman, B. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 2004, 78, 1440–1447. [Google Scholar] [CrossRef] [Green Version]
- Kotenko, S.V.; Saccani, S.; Izotova, L.S.; Mirochnitchenko, O.V.; Pestka, S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 2000, 97, 1695–1700. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, C.; Garcia, W.; Godwin, M.J.; Spencer, J.V.; Stern, J.L.; Abendroth, A.; Slobedman, B. Immunomodulatory properties of a viral homolog of human interleukin-10 expressed by human cytomegalovirus during the latent phase of infection. J. Virol. 2008, 82, 3736–3750. [Google Scholar] [CrossRef] [Green Version]
- Cheung, A.K.; Gottlieb, D.J.; Plachter, B.; Pepperl-Klindworth, S.; Avdic, S.; Cunningham, A.L.; Abendroth, A.; Slobedman, B. The role of the human cytomegalovirus UL111A gene in down-regulating CD4+ T-cell recognition of latently infected cells: Implications for virus elimination during latency. Blood 2009, 114, 4128–4137. [Google Scholar] [CrossRef] [Green Version]
- Reeves, M.B.; Davies, A.A.; McSharry, B.P.; Wilkinson, G.W.; Sinclair, J.H. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 2007, 316, 1345–1348. [Google Scholar] [CrossRef] [Green Version]
- Poole, E.; Walther, A.; Raven, K.; Benedict, C.A.; Mason, G.M.; Sinclair, J. The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner. J. Virol. 2013, 87, 4261–4271. [Google Scholar] [CrossRef] [Green Version]
- Benedict, C.A.; Butrovich, K.D.; Lurain, N.S.; Corbeil, J.; Rooney, I.; Schneider, P.; Tschopp, J.; Ware, C.F. Cutting edge: A novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J. Immunol. 1999, 162, 6967–6970. [Google Scholar]
- Rossetto, C.C.; Tarrant-Elorza, M.; Pari, G.S. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013, 9, e1003366. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Caviness, K.; Buehler, J.; Smithey, M.; Nikolich-Žugich, J.; Goodrum, F. Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc. Natl. Acad. Sci. USA 2017, 114, E10586–E10595. [Google Scholar] [CrossRef] [Green Version]
- Shnayder, M.; Nachshon, A.; Krishna, B.; Poole, E.; Boshkov, A.; Binyamin, A.; Maza, I.; Sinclair, J.; Schwartz, M.; Stern-Ginossar, N. Defining the Transcriptional Landscape during Cytomegalovirus Latency with Single-Cell RNA Sequencing. MBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Grinde, B. Herpesviruses: Latency and reactivation—Viral strategies and host response. J. Oral. Microbiol. 2013, 5. [Google Scholar] [CrossRef] [Green Version]
- Collin, V.; Flamand, L. HHV-6A/B Integration and the Pathogenesis Associated with the Reactivation of Chromosomally Integrated HHV-6A/B. Viruses 2017, 9, 160. [Google Scholar] [CrossRef] [Green Version]
- Kaufer, B.B.; Flamand, L. Chromosomally integrated HHV-6: Impact on virus, cell and organismal biology. Curr Opin Virol 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Finkel, Y.; Schmiedel, D.; Tai-Schmiedel, J.; Nachshon, A.; Winkler, R.; Dobesova, M.; Schwartz, M.; Mandelboim, O.; Stern-Ginossar, N. Comprehensive annotations of human herpesvirus 6A and 6B genomes reveal novel and conserved genomic features. Elife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.; Shimada, K.; Sashihara, J.; Tanaka-Taya, K.; Yamanishi, K. Identification of human herpesvirus 6 latency-associated transcripts. J. Virol. 2002, 76, 4145–4151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, K.; Sashihara, J.; Shimada, K.; Takemoto, M.; Amo, K.; Miyagawa, H.; Yamanishi, K. Recognition of a novel stage of betaherpesvirus latency in human herpesvirus 6. J. Virol. 2003, 77, 2258–2264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotola, A.; Ravaioli, T.; Gonelli, A.; Dewhurst, S.; Cassai, E.; Di Luca, D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc. Natl. Acad. Sci. USA 1998, 95, 13911–13916. [Google Scholar] [CrossRef] [Green Version]
- Caselli, E.; Bracci, A.; Galvan, M.; Boni, M.; Rotola, A.; Bergamini, C.; Cermelli, C.; Dal Monte, P.; Gompels, U.A.; Cassai, E.; et al. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology 2006, 346, 402–414. [Google Scholar] [CrossRef] [Green Version]
- Saviola, A.J.; Zimmermann, C.; Mariani, M.P.; Signorelli, S.A.; Gerrard, D.L.; Boyd, J.R.; Wight, D.J.; Morissette, G.; Gravel, A.; Dubuc, I.; et al. Chromatin Profiles of Chromosomally Integrated Human Herpesvirus-6A. Front. Microbiol. 2019, 10, 1408. [Google Scholar] [CrossRef]
- Piedade, D.; Azevedo-Pereira, J.M. The Role of microRNAs in the Pathogenesis of Herpesvirus Infection. Viruses 2016, 8, 156. [Google Scholar] [CrossRef] [Green Version]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef] [Green Version]
- Prusty, B.K.; Gulve, N.; Chowdhury, S.R.; Schuster, M.; Strempel, S.; Descamps, V.; Rudel, T. HHV-6 encoded small non-coding RNAs define an intermediate and early stage in viral reactivation. NPJ Genom. Med. 2018, 3, 25. [Google Scholar] [CrossRef]
- Nukui, M.; Mori, Y.; Murphy, E.A. A human herpesvirus 6A-encoded microRNA: Role in viral lytic replication. J. Virol. 2015, 89, 2615–2627. [Google Scholar] [CrossRef] [Green Version]
- Stevens, J.G.; Wagner, E.K.; Devi-Rao, G.B.; Cook, M.L.; Feldman, L.T. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 1987, 235, 1056–1059. [Google Scholar] [CrossRef]
- Wagner, E.K.; Bloom, D.C. Experimental investigation of herpes simplex virus latency. Clin. Microbiol. Rev. 1997, 10, 419–443. [Google Scholar] [CrossRef]
- Depledge, D.P.; Sadaoka, T.; Ouwendijk, W.J.D. Molecular Aspects of Varicella-Zoster Virus Latency. Viruses 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Javier, R.T.; Stevens, J.G.; Dissette, V.B.; Wagner, E.K. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology 1988, 166, 254–257. [Google Scholar] [CrossRef]
- Sedarati, F.; Izumi, K.M.; Wagner, E.K.; Stevens, J.G. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. J. Virol. 1989, 63, 4455–4458. [Google Scholar] [CrossRef] [Green Version]
- Steiner, I.; Spivack, J.G.; Lirette, R.P.; Brown, S.M.; MacLean, A.R.; Subak-Sharpe, J.H.; Fraser, N.W. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 1989, 8, 505–511. [Google Scholar] [CrossRef]
- Amelio, A.L.; Giordani, N.V.; Kubat, N.J.; O’neil, J.E.; Bloom, D.C. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 2006, 80, 2063–2068. [Google Scholar] [CrossRef] [Green Version]
- Cliffe, A.R.; Garber, D.A.; Knipe, D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009, 83, 8182–8190. [Google Scholar] [CrossRef] [Green Version]
- Kwiatkowski, D.L.; Thompson, H.W.; Bloom, D.C. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 2009, 83, 8173–8181. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.Y.; Zhou, C.; Johnson, K.E.; Colgrove, R.C.; Coen, D.M.; Knipe, D.M. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 2005, 102, 16055–16059. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.; Lock, M.; Miller, C.G.; Fraser, N.W. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J. Virol. 2002, 76, 717–729. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Chentoufi, A.A.; Hsiang, C.; Carpenter, D.; Osorio, N.; BenMohamed, L.; Fraser, N.W.; Jones, C.; Wechsler, S.L. The herpes simplex virus type 1 latency-associated transcript can protect neuron-derived C1300 and Neuro2A cells from granzyme B-induced apoptosis and CD8 T-cell killing. J. Virol. 2011, 85, 2325–2332. [Google Scholar] [CrossRef] [Green Version]
- Perng, G.C.; Jones, C.; Ciacci-Zanella, J.; Stone, M.; Henderson, G.; Yukht, A.; Slanina, S.M.; Hofman, F.M.; Ghiasi, H.; Nesburn, A.B.; et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 2000, 287, 1500–1503. [Google Scholar] [CrossRef] [Green Version]
- Held, K.; Junker, A.; Dornmair, K.; Meinl, E.; Sinicina, I.; Brandt, T.; Theil, D.; Derfuss, T. Expression of herpes simplex virus 1-encoded microRNAs in human trigeminal ganglia and their relation to local T-cell infiltrates. J. Virol. 2011, 85, 9680–9685. [Google Scholar] [CrossRef] [Green Version]
- Jurak, I.; Kramer, M.F.; Mellor, J.C.; van Lint, A.L.; Roth, F.P.; Knipe, D.M.; Coen, D.M. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J. Virol. 2010, 84, 4659–4672. [Google Scholar] [CrossRef] [Green Version]
- Kramer, M.F.; Jurak, I.; Pesola, J.M.; Boissel, S.; Knipe, D.M.; Coen, D.M. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology 2011, 417, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef] [Green Version]
- Flores, O.; Nakayama, S.; Whisnant, A.W.; Javanbakht, H.; Cullen, B.R.; Bloom, D.C. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J. Virol. 2013, 87, 6589–6603. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.H.; Lee, L.Y.; Garber, D.A.; Schaffer, P.A.; Knipe, D.M.; Coen, D.M. Neither LAT nor open reading frame P mutations increase expression of spliced or intron-containing ICP0 transcripts in mouse ganglia latently infected with herpes simplex virus. J. Virol. 2002, 76, 4764–4772. [Google Scholar] [CrossRef] [Green Version]
- Feldman, L.T.; Ellison, A.R.; Voytek, C.C.; Yang, L.; Krause, P.; Margolis, T.P. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proc. Natl. Acad. Sci. USA 2002, 99, 978–983. [Google Scholar] [CrossRef] [Green Version]
- Giordani, N.V.; Neumann, D.M.; Kwiatkowski, D.L.; Bhattacharjee, P.S.; McAnany, P.K.; Hill, J.M.; Bloom, D.C. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J. Virol. 2008, 82, 6056–6060. [Google Scholar] [CrossRef] [Green Version]
- Green, M.T.; Rosborough, J.P.; Dunkel, E.C. In vivo reactivation of herpes simplex virus in rabbit trigeminal ganglia: Electrode model. Infect. Immun. 1981, 34, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Kramer, M.F.; Coen, D.M. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 1995, 69, 1389–1399. [Google Scholar] [CrossRef] [Green Version]
- Kramer, M.F.; Chen, S.H.; Knipe, D.M.; Coen, D.M. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J. Virol. 1998, 72, 1177–1185. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Raja, P.; Pan, D.; Pesola, J.M.; Coen, D.M.; Knipe, D.M. CCCTC-Binding Factor Acts as a Heterochromatin Barrier on Herpes Simplex Viral Latent Chromatin and Contributes to Poised Latent Infection. MBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Maillet, S.; Naas, T.; Crepin, S.; Roque-Afonso, A.M.; Lafay, F.; Efstathiou, S.; Labetoulle, M. Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts. J. Virol. 2006, 80, 9310–9321. [Google Scholar] [CrossRef] [Green Version]
- Nicoll, M.P.; Hann, W.; Shivkumar, M.; Harman, L.E.; Connor, V.; Coleman, H.M.; Proença, J.T.; Efstathiou, S. The HSV-1 Latency-Associated Transcript Functions to Repress Latent Phase Lytic Gene Expression and Suppress Virus Reactivation from Latently Infected Neurons. PLoS Pathog. 2016, 12, e1005539. [Google Scholar] [CrossRef]
- Pesola, J.M.; Zhu, J.; Knipe, D.M.; Coen, D.M. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J. Virol. 2005, 79, 14516–14525. [Google Scholar] [CrossRef] [Green Version]
- Raja, P.; Lee, J.S.; Pan, D.; Pesola, J.M.; Coen, D.M.; Knipe, D.M. A Herpesviral Lytic Protein Regulates the Structure of Latent Viral Chromatin. MBio 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Russell, T.A.; Tscharke, D.C. Lytic Promoters Express Protein during Herpes Simplex Virus Latency. PLoS Pathog. 2016, 12, e1005729. [Google Scholar] [CrossRef]
- Tal-Singer, R.; Lasner, T.M.; Podrzucki, W.; Skokotas, A.; Leary, J.J.; Berger, S.L.; Fraser, N.W. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J. Virol. 1997, 71, 5268–5276. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Tscharke, D.C. Herpes simplex virus latency is noisier the closer we look. J. Virol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.S.; Danaher, R.J. Asymptomatic shedding of herpes simplex virus (HSV) in the oral cavity. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2008, 105, 43–50. [Google Scholar] [CrossRef]
- Ramchandani, M.S.; Jing, L.; Russell, R.M.; Tran, T.; Laing, K.J.; Magaret, A.S.; Selke, S.; Cheng, A.; Huang, M.L.; Xie, H.; et al. Viral Genetics Modulate Orolabial Herpes Simplex Virus Type 1 Shedding in Humans. J. Infect. Dis. 2019, 219, 1058–1066. [Google Scholar] [CrossRef]
- Chen, X.P.; Mata, M.; Kelley, M.; Glorioso, J.C.; Fink, D.J. The relationship of herpes simplex virus latency associated transcript expression to genome copy number: A quantitative study using laser capture microdissection. J. Neurovirol. 2002, 8, 204–210. [Google Scholar] [CrossRef]
- Maggioncalda, J.; Mehta, A.; Su, Y.H.; Fraser, N.W.; Block, T.M. Correlation between herpes simplex virus type 1 rate of reactivation from latent infection and the number of infected neurons in trigeminal ganglia. Virology 1996, 225, 72–81. [Google Scholar] [CrossRef] [Green Version]
- Mehta, A.; Maggioncalda, J.; Bagasra, O.; Thikkavarapu, S.; Saikumari, P.; Valyi-Nagy, T.; Fraser, N.W.; Block, T.M. In situ DNA PCR and RNA hybridization detection of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology 1995, 206, 633–640. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Lau, T.Y.; Morales, M.; Mont, E.K.; Straus, S.E. Laser-capture microdissection: Refining estimates of the quantity and distribution of latent herpes simplex virus 1 and varicella-zoster virus DNA in human trigeminal Ganglia at the single-cell level. J. Virol. 2005, 79, 14079–14087. [Google Scholar] [CrossRef] [Green Version]
- Cliffe, A.R.; Wilson, A.C. Restarting Lytic Gene Transcription at the Onset of Herpes Simplex Virus Reactivation. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Bloom, D.C. Alphaherpesvirus Latency: A Dynamic State of Transcription and Reactivation. Adv. Virus Res. 2016, 94, 53–80. [Google Scholar] [CrossRef]
- Nicoll, M.P.; Proença, J.T.; Efstathiou, S. The molecular basis of herpes simplex virus latency. FEMS Microbiol. Rev. 2012, 36, 684–705. [Google Scholar] [CrossRef]
- Cohrs, R.J.; Barbour, M.; Gilden, D.H. Varicella-zoster virus (VZV) transcription during latency in human ganglia: Detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 1996, 70, 2789–2796. [Google Scholar] [CrossRef] [Green Version]
- Cohrs, R.J.; Gilden, D.H.; Kinchington, P.R.; Grinfeld, E.; Kennedy, P.G. Varicella-zoster virus gene 66 transcription and translation in latently infected human Ganglia. J. Virol. 2003, 77, 6660–6665. [Google Scholar] [CrossRef] [Green Version]
- Cohrs, R.J.; Gilden, D.H. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J. Virol. 2007, 81, 2950–2956. [Google Scholar] [CrossRef] [Green Version]
- Mahalingam, R.; Wellish, M.; Cohrs, R.; Debrus, S.; Piette, J.; Rentier, B.; Gilden, D.H. Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc. Natl. Acad. Sci. USA 1996, 93, 2122–2124. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.I.; Cox, E.; Pesnicak, L.; Srinivas, S.; Krogmann, T. The varicella-zoster virus open reading frame 63 latency-associated protein is critical for establishment of latency. J. Virol. 2004, 78, 11833–11840. [Google Scholar] [CrossRef] [Green Version]
- Hood, C.; Cunningham, A.L.; Slobedman, B.; Arvin, A.M.; Sommer, M.H.; Kinchington, P.R.; Abendroth, A. Varicella-zoster virus ORF63 inhibits apoptosis of primary human neurons. J. Virol. 2006, 80, 1025–1031. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.I.; Krogmann, T.; Ross, J.P.; Pesnicak, L.; Prikhod’ko, E.A. Varicella-zoster virus ORF4 latency-associated protein is important for establishment of latency. J. Virol. 2005, 79, 6969–6975. [Google Scholar] [CrossRef] [Green Version]
- Depledge, D.P.; Ouwendijk, W.J.D.; Sadaoka, T.; Braspenning, S.E.; Mori, Y.; Cohrs, R.J.; Verjans, G.M.G.M.; Breuer, J. A spliced latency-associated VZV transcript maps antisense to the viral transactivator gene 61. Nat. Commun. 2018, 9, 1167. [Google Scholar] [CrossRef]
- Sadaoka, T.; Rajbhandari, L.; Shukla, P.; Jagdish, B.; Lee, H.; Lee, G.; Venkatesan, A. Human stem cell derived sensory neurons are positioned to support varicella zoster virus latency. bioRxiv 2020. [Google Scholar] [CrossRef]
- Tang, Q.; Qin, D.; Lv, Z.; Zhu, X.; Ma, X.; Yan, Q.; Zeng, Y.; Guo, Y.; Feng, N.; Lu, C. Herpes simplex virus type 2 triggers reactivation of Kaposi’s sarcoma-associated herpesvirus from latency and collaborates with HIV-1 Tat. PLoS ONE 2012, 7, e31652. [Google Scholar] [CrossRef]
- Ye, F.; Lei, X.; Gao, S.J. Mechanisms of Kaposi’s Sarcoma-Associated Herpesvirus Latency and Reactivation. Adv. Virol. 2011, 2011. [Google Scholar] [CrossRef]
- Purushothaman, P.; Uppal, T.; Verma, S.C. Molecular biology of KSHV lytic reactivation. Viruses 2015, 7, 116–153. [Google Scholar] [CrossRef] [Green Version]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef]
- Flamand, L.; Stefanescu, I.; Ablashi, D.V.; Menezes, J. Activation of the Epstein–Barr virus replicative cycle by human herpesvirus 6. J. Virol. 1993, 67, 6768–6777. [Google Scholar] [CrossRef] [Green Version]
- Reynaldi, A.; Schlub, T.E.; Chelimo, K.; Sumba, P.O.; Piriou, E.; Ogolla, S.; Moormann, A.M.; Rochford, R.; Davenport, M.P. Impact of Plasmodium falciparum Coinfection on Longitudinal Epstein–Barr Virus Kinetics in Kenyan Children. J. Infect. Dis. 2016, 213, 985–991. [Google Scholar] [CrossRef] [Green Version]
- Karrasch, M.; Herfurth, K.; Kläver, M.; Miethke, J.; Mayer-Scholl, A.; Luge, E.; Straube, E.; Busch, M. Severe leptospirosis complicated by Epstein–Barr Virus reactivation. Infection 2015, 43, 763–769. [Google Scholar] [CrossRef]
- Ueda, S.; Uchiyama, S.; Azzi, T.; Gysin, C.; Berger, C.; Bernasconi, M.; Harabuchi, Y.; Zinkernagel, A.S.; Nadal, D. Oropharyngeal group A streptococcal colonization disrupts latent Epstein–Barr virus infection. J. Infect. Dis. 2014, 209, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Byrne, C.M.; Johnston, C.; Orem, J.; Okuku, F.; Huang, M.-L.; Selke, S.; Wald, A.; Corey, L.; Schiffer, J.; Casper, T.C.; et al. Increased oral Epstein–Barr virus shedding with HIV-1 co-infection is due to a combination of B cell activation and impaired cellular immune control. bioRxiv 2019. [Google Scholar] [CrossRef]
- Makielski, K.R.; Lee, D.; Lorenz, L.D.; Nawandar, D.M.; Chiu, Y.F.; Kenney, S.C.; Lambert, P.F. Human papillomavirus promotes Epstein–Barr virus maintenance and lytic reactivation in immortalized oral keratinocytes. Virology 2016, 495, 52–62. [Google Scholar] [CrossRef]
- Hirsiger, J.R.; Fuchs, P.S.; Häusermann, P.; Müller-Durovic, B.; Daikeler, T.; Recher, M.; Hirsch, H.H.; Terracciano, L.; Berger, C.T. Syphilis Reactivates Latent Epstein–Barr Virus Reservoir via Toll-Like Receptor 2 and B-Cell Receptor Activation. Open Forum Infect. Dis. 2019, 6, ofz317. [Google Scholar] [CrossRef]
- Murata, T.; Tsurumi, T. Switching of EBV cycles between latent and lytic states. Rev. Med. Virol. 2014, 24, 142–153. [Google Scholar] [CrossRef]
- Mercader, M.; Taddeo, B.; Panella, J.R.; Chandran, B.; Nickoloff, B.J.; Foreman, K.E. Induction of HHV-8 lytic cycle replication by inflammatory cytokines produced by HIV-1-infected T cells. Am. J. Pathol. 2000, 156, 1961–1971. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Renne, R.; Dittmer, D.; Ganem, D. Inflammatory cytokines and the reactivation of Kaposi’s sarcoma-associated herpesvirus lytic replication. Virology 2000, 266, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Blackbourn, D.J.; Fujimura, S.; Kutzkey, T.; Levy, J.A. Induction of human herpesvirus-8 gene expression by recombinant interferon gamma. AIDS 2000, 14, 98–99. [Google Scholar] [CrossRef]
- Gandhi, J.; Gaur, N.; Khera, L.; Kaul, R.; Robertson, E.S. COX-2 induces lytic reactivation of EBV through PGE2 by modulating the EP receptor signaling pathway. Virology 2015, 484, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Davis, D.A.; Rinderknecht, A.S.; Zoeteweij, J.P.; Aoki, Y.; Read-Connole, E.L.; Tosato, G.; Blauvelt, A.; Yarchoan, R. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 2001, 97, 3244–3250. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.H.; Wang, N.; Li, A.; Liao, W.T.; Pan, Z.G.; Mai, S.J.; Li, D.J.; Zeng, M.S.; Wen, J.M.; Zeng, Y.X. Hypoxia can contribute to the induction of the Epstein–Barr virus (EBV) lytic cycle. J. Clin. Virol. 2006, 37, 98–103. [Google Scholar] [CrossRef]
- Kraus, R.J.; Yu, X.; Cordes, B.A.; Sathiamoorthi, S.; Iempridee, T.; Nawandar, D.M.; Ma, S.; Romero-Masters, J.C.; McChesney, K.G.; Lin, Z.; et al. Hypoxia-inducible factor-1α plays roles in Epstein–Barr virus’s natural life cycle and tumorigenesis by inducing lytic infection through direct binding to the immediate-early BZLF1 gene promoter. PLoS Pathog. 2017, 13, e1006404. [Google Scholar] [CrossRef]
- Ye, F.; Zhou, F.; Bedolla, R.G.; Jones, T.; Lei, X.; Kang, T.; Guadalupe, M.; Gao, S.J. Reactive oxygen species hydrogen peroxide mediates Kaposi’s sarcoma-associated herpesvirus reactivation from latency. PLoS Pathog. 2011, 7, e1002054. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Feng, J.; Sun, R. Oxidative stress induces reactivation of Kaposi’s sarcoma-associated herpesvirus and death of primary effusion lymphoma cells. J. Virol. 2011, 85, 715–724. [Google Scholar] [CrossRef] [Green Version]
- Coskun, O.; Sener, K.; Kilic, S.; Erdem, H.; Yaman, H.; Besirbellioglu, A.B.; Gul, H.C.; Eyigun, C.P. Stress-related Epstein–Barr virus reactivation. Clin. Exp. Med. 2010, 10, 15–20. [Google Scholar] [CrossRef]
- Mehta, S.K.; Laudenslager, M.L.; Stowe, R.P.; Crucian, B.E.; Sams, C.F.; Pierson, D.L. Multiple latent viruses reactivate in astronauts during Space Shuttle missions. Brain Behav. Immun. 2014, 41, 210–217. [Google Scholar] [CrossRef]
- Mehta, S.K.; Laudenslager, M.L.; Stowe, R.P.; Crucian, B.E.; Feiveson, A.H.; Sams, C.F.; Pierson, D.L. Latent virus reactivation in astronauts on the international space station. NPJ Microgravity 2017, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Pierson, D.L.; Stowe, R.P.; Phillips, T.M.; Lugg, D.J.; Mehta, S.K. Epstein–Barr virus shedding by astronauts during space flight. Brain Behav. Immun. 2005, 19, 235–242. [Google Scholar] [CrossRef]
- Rooney, B.V.; Crucian, B.E.; Pierson, D.L.; Laudenslager, M.L.; Mehta, S.K. Herpes Virus Reactivation in Astronauts during Spaceflight and Its Application on Earth. Front. Microbiol. 2019, 10, 16. [Google Scholar] [CrossRef]
- Feng, W.H.; Israel, B.; Raab-Traub, N.; Busson, P.; Kenney, S.C. Chemotherapy induces lytic EBV replication and confers ganciclovir susceptibility to EBV-positive epithelial cell tumors. Cancer Res. 2002, 62, 1920–1926. [Google Scholar]
- Westphal, E.M.; Blackstock, W.; Feng, W.; Israel, B.; Kenney, S.C. Activation of lytic Epstein–Barr virus (EBV) infection by radiation and sodium butyrate in vitro and in vivo: A potential method for treating EBV-positive malignancies. Cancer Res. 2000, 60, 5781–5788. [Google Scholar]
- Davies, A.H.; Grand, R.J.; Evans, F.J.; Rickinson, A.B. Induction of Epstein–Barr virus lytic cycle by tumor-promoting and non-tumor-promoting phorbol esters requires active protein kinase C. J. Virol. 1991, 65, 6838–6844. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Ikuta, K.; Tajima, M.; Sairenji, T. 12-O-tetradecanoylphorbol-13-acetate induces Epstein–Barr virus reactivation via NF-kappaB and AP-1 as regulated by protein kinase C and mitogen-activated protein kinase. Virology 2001, 286, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Gorres, K.L.; Daigle, D.; Mohanram, S.; Miller, G. Activation and repression of Epstein–Barr Virus and Kaposi’s sarcoma-associated herpesvirus lytic cycles by short- and medium-chain fatty acids. J. Virol. 2014, 88, 8028–8044. [Google Scholar] [CrossRef] [Green Version]
- Renne, R.; Zhong, W.; Herndier, B.; McGrath, M.; Abbey, N.; Kedes, D.; Ganem, D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996, 2, 342–346. [Google Scholar] [CrossRef]
- Guito, J.; Lukac, D.M. KSHV Rta Promoter Specification and Viral Reactivation. Front. Microbiol. 2012, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Reddehase, M.J.; Lemmermann, N.A.W. Cellular reservoirs of latent cytomegaloviruses. Med. Microbiol. Immunol. 2019, 208, 391–403. [Google Scholar] [CrossRef]
- Zhu, D.; Pan, C.; Sheng, J.; Liang, H.; Bian, Z.; Liu, Y.; Trang, P.; Wu, J.; Liu, F.; Zhang, C.Y.; et al. Human cytomegalovirus reprograms haematopoietic progenitor cells into immunosuppressive monocytes to achieve latency. Nat. Microbiol. 2018, 3, 503–513. [Google Scholar] [CrossRef]
- Söderberg-Nauclér, C.; Fish, K.N.; Nelson, J.A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 1997, 91, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Lautenschlager, I.; Lappalainen, M.; Linnavuori, K.; Suni, J.; Höckerstedt, K. CMV infection is usually associated with concurrent HHV-6 and HHV-7 antigenemia in liver transplant patients. J. Clin. Virol. 2002, 25 (Suppl. 2), S57–S61. [Google Scholar] [CrossRef]
- Gerdemann, U.; Keukens, L.; Keirnan, J.M.; Katari, U.L.; Nguyen, C.T.; de Pagter, A.P.; Ramos, C.A.; Kennedy-Nasser, A.; Gottschalk, S.M.; Heslop, H.E.; et al. Immunotherapeutic strategies to prevent and treat human herpesvirus 6 reactivation after allogeneic stem cell transplantation. Blood 2013, 121, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Mendez, J.C.; Dockrell, D.H.; Espy, M.J.; Smith, T.F.; Wilson, J.A.; Harmsen, W.S.; Ilstrup, D.; Paya, C.V. Human beta-herpesvirus interactions in solid organ transplant recipients. J. Infect. Dis. 2001, 183, 179–184. [Google Scholar] [CrossRef]
- Fan, J.; Jing, M.; Yang, M.; Xu, L.; Liang, H.; Huang, Y.; Yang, R.; Gui, G.; Wang, H.; Gong, S.; et al. Herpesvirus infections in hematopoietic stem cell transplant recipients seropositive for human cytomegalovirus before transplantation. Int. J. Infect. Dis. 2016, 46, 89–93. [Google Scholar] [CrossRef] [Green Version]
- Shams El-Din, A.A.; El-Desoukey, N.A.; Amin Tawadrous, D.G.; Fouad, N.M.B.E.; Abdel-Mooti, M.; Hotar, S.F. The potential association of CMV-specific CD8+ T lymphocyte reconstitution with the risk of CMV reactivation and persistency in post allogeneic stem cell transplant patients. Hematology 2018, 23, 463–469. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, J.; Sissons, P. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 2006, 87, 1763–1779. [Google Scholar] [CrossRef]
- Chapenko, S.; Folkmane, I.; Tomsone, V.; Amerika, D.; Rozentals, R.; Murovska, M. Co-infection of two beta-herpesviruses (CMV and HHV-7) as an increased risk factor for ‘CMV disease’ in patients undergoing renal transplantation. Clin. Transplant. 2000, 14, 486–492. [Google Scholar] [CrossRef]
- Mehta, S.K.; Stowe, R.P.; Feiveson, A.H.; Tyring, S.K.; Pierson, D.L. Reactivation and shedding of cytomegalovirus in astronauts during spaceflight. J. Infect. Dis. 2000, 182, 1761–1764. [Google Scholar] [CrossRef] [Green Version]
- D’Aiuto, L.; Di Maio, R.; Heath, B.; Raimondi, G.; Milosevic, J.; Watson, A.M.; Bamne, M.; Parks, W.T.; Yang, L.; Lin, B.; et al. Human induced pluripotent stem cell-derived models to investigate human cytomegalovirus infection in neural cells. PLoS ONE 2012, 7, e49700. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.H.; Schwartz, P.H.; Fortunato, E.A. Neonatal neural progenitor cells and their neuronal and glial cell derivatives are fully permissive for human cytomegalovirus infection. J. Virol. 2008, 82, 9994–10007. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.H.; Hannemann, H.; Kulkarni, A.S.; Schwartz, P.H.; O’Dowd, J.M.; Fortunato, E.A. Human cytomegalovirus infection causes premature and abnormal differentiation of human neural progenitor cells. J. Virol. 2010, 84, 3528–3541. [Google Scholar] [CrossRef] [Green Version]
- Odeberg, J.; Wolmer, N.; Falci, S.; Westgren, M.; Seiger, A.; Söderberg-Nauclér, C. Human cytomegalovirus inhibits neuronal differentiation and induces apoptosis in human neural precursor cells. J. Virol. 2006, 80, 8929–8939. [Google Scholar] [CrossRef] [Green Version]
- Taylor-Wiedeman, J.; Sissons, P.; Sinclair, J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 1994, 68, 1597–1604. [Google Scholar] [CrossRef] [Green Version]
- Reeves, M.B.; MacAry, P.A.; Lehner, P.J.; Sissons, J.G.; Sinclair, J.H. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 2005, 102, 4140–4145. [Google Scholar] [CrossRef] [Green Version]
- O’Connor, C.M.; Murphy, E.A. A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J. Virol. 2012, 86, 9854–9865. [Google Scholar] [CrossRef] [Green Version]
- Yasukawa, M.; Ohminami, H.; Sada, E.; Yakushijin, Y.; Kaneko, M.; Yanagisawa, K.; Kohno, H.; Bando, S.; Fujita, S. Latent infection and reactivation of human herpesvirus 6 in two novel myeloid cell lines. Blood 1999, 93, 991–999. [Google Scholar] [CrossRef]
- Ahlqvist, J.; Fotheringham, J.; Akhyani, N.; Yao, K.; Fogdell-Hahn, A.; Jacobson, S. Differential tropism of human herpesvirus 6 (HHV-6) variants and induction of latency by HHV-6A in oligodendrocytes. J. Neurovirol. 2005, 11, 384–394. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Asano, Y.; Akimoto, S.; Ozaki, T.; Iwasaki, T.; Kurata, T.; Goshima, F.; Nishiyama, Y. Latent infection of human herpesvirus 6 in astrocytoma cell line and alteration of cytokine synthesis. J. Med. Virol. 2002, 66, 497–505. [Google Scholar] [CrossRef]
- Katsafanas, G.C.; Schirmer, E.C.; Wyatt, L.S.; Frenkel, N. In vitro activation of human herpesviruses 6 and 7 from latency. Proc. Natl. Acad. Sci. USA 1996, 93, 9788–9792. [Google Scholar] [CrossRef] [Green Version]
- Kondo, K.; Kondo, T.; Okuno, T.; Takahashi, M.; Yamanishi, K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 1991, 72 Pt 6, 1401–1408. [Google Scholar] [CrossRef]
- Prasad, A.; Remick, J.; Zeichner, S.L. Activation of human herpesvirus replication by apoptosis. J. Virol. 2013, 87, 10641–10650. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, P.G.; Rovnak, J.; Badani, H.; Cohrs, R.J. A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation. J. Gen. Virol. 2015, 96, 1581–1602. [Google Scholar] [CrossRef]
- Wilson, A.C.; Mohr, I. A cultured affair: HSV latency and reactivation in neurons. Trends Microbiol 2012, 20, 604–611. [Google Scholar] [CrossRef] [Green Version]
- Mazzarello, V.; Ferrari, M.; Decandia, S.; Sotgiu, M.A. Sunlight and Herpes Virus. In Human Herpesvirus Infection—Biological Features, Transmission, Symptoms, Diagnosis and Treatment; Thomasini, R.L., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Zak-Prelich, M.; Borkowski, J.L.; Alexander, F.; Norval, M. The role of solar ultraviolet irradiation in zoster. Epidemiol Infect 2002, 129, 593–597. [Google Scholar] [CrossRef]
- Nussbaum, R. Theories on Varicella Zoster Virus Reactivation Based on Shingles Patterns. Sci. J. Lander Coll. Arts Sci. 2014, 8, 10. [Google Scholar]
- Fatahzadeh, M.; Schwartz, R.A. Human herpes simplex virus infections: Epidemiology, pathogenesis, symptomatology, diagnosis, and management. J. Am. Acad. Dermatol. 2007, 57, 737–763. [Google Scholar] [CrossRef]
- Black, B.J.; Atmaramani, R.; Kumaraju, R.; Plagens, S.; Romero-Ortega, M.; Dussor, G.; Price, T.J.; Campbell, Z.T.; Pancrazio, J.J. Adult mouse sensory neurons on microelectrode arrays exhibit increased spontaneous and stimulus-evoked activity in the presence of interleukin-6. J. Neurophysiol. 2018, 120, 1374–1385. [Google Scholar] [CrossRef]
- Poon, D.C.; Ho, Y.S.; Chiu, K.; Wong, H.L.; Chang, R.C. Sickness: From the focus on cytokines, prostaglandins, and complement factors to the perspectives of neurons. Neurosci. Biobehav. Rev. 2015, 57, 30–45. [Google Scholar] [CrossRef]
- Harbour, D.A.; Blyth, W.A.; Hill, T.J. Prostanglandins enhance spread of herpes simplex virus in cell cultures. J. Gen. Virol. 1978, 41, 87–95. [Google Scholar] [CrossRef]
- Freeman, M.L.; Sheridan, B.S.; Bonneau, R.H.; Hendricks, R.L. Psychological stress compromises CD8+ T cell control of latent herpes simplex virus type 1 infections. J. Immunol. 2007, 179, 322–328. [Google Scholar] [CrossRef] [Green Version]
- Schmader, K.; Studenski, S.; MacMillan, J.; Grufferman, S.; Cohen, H.J. Are stressful life events risk factors for herpes zoster? J. Am. Geriatr. Soc. 1990, 38, 1188–1194. [Google Scholar] [CrossRef]
- Schmader, K.; George, L.K.; Burchett, B.M.; Hamilton, J.D.; Pieper, C.F. Race and stress in the incidence of herpes zoster in older adults. J. Am. Geriatr. Soc. 1998, 46, 973–977. [Google Scholar] [CrossRef]
- Lasserre, A.; Blaizeau, F.; Gorwood, P.; Bloch, K.; Chauvin, P.; Liard, F.; Blanchon, T.; Hanslik, T. Herpes zoster: Family history and psychological stress-case-control study. J. Clin. Virol. 2012, 55, 153–157. [Google Scholar] [CrossRef]
- Hsia, S.C.; Bedadala, G.R.; Balish, M.D. Effects of thyroid hormone on HSV-1 gene regulation: Implications in the control of viral latency and reactivation. Cell Biosci. 2011, 1, 24. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Wilson, A.C.; Chao, M.V.; Mohr, I. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev. 2012, 26, 1527–1532. [Google Scholar] [CrossRef] [Green Version]
- Yanez, A.A.; Harrell, T.; Sriranganathan, H.J.; Ives, A.M.; Bertke, A.S. Neurotrophic Factors NGF, GDNF and NTN Selectively Modulate HSV1 and HSV2 Lytic Infection and Reactivation in Primary Adult Sensory and Autonomic Neurons. Pathogens 2017, 6, 5. [Google Scholar] [CrossRef]
- Camarena, V.; Kobayashi, M.; Kim, J.Y.; Roehm, P.; Perez, R.; Gardner, J.; Wilson, A.C.; Mohr, I.; Chao, M.V. Nature and duration of growth factor signaling through receptor tyrosine kinases regulates HSV-1 latency in neurons. Cell Host Microbe 2010, 8, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Wilcox, C.L.; Johnson, E.M. Nerve growth factor deprivation results in the reactivation of latent herpes simplex virus in vitro. J. Virol. 1987, 61, 2311–2315. [Google Scholar] [CrossRef] [Green Version]
- Cohrs, R.J.; Badani, H.; Baird, N.L.; White, T.M.; Sanford, B.; Gilden, D. Induction of varicella zoster virus DNA replication in dissociated human trigeminal ganglia. J. Neurovirol. 2017, 23, 152–157. [Google Scholar] [CrossRef] [Green Version]
- Pourchet, A.; Modrek, A.S.; Placantonakis, D.G.; Mohr, I.; Wilson, A.C. Modeling HSV-1 Latency in Human Embryonic Stem Cell-Derived Neurons. Pathogens 2017, 6, 24. [Google Scholar] [CrossRef] [Green Version]
- Laemmle, L.; Goldstein, R.S.; Kinchington, P.R. Modeling Varicella Zoster Virus Persistence and Reactivation—Closer to Resolving a Perplexing Persistent State. Front. Microbiol. 2019, 10, 1634. [Google Scholar] [CrossRef] [Green Version]
- Cliffe, A.R.; Arbuckle, J.H.; Vogel, J.L.; Geden, M.J.; Rothbart, S.B.; Cusack, C.L.; Strahl, B.D.; Kristie, T.M.; Deshmukh, M. Neuronal Stress Pathway Mediating a Histone Methyl/Phospho Switch Is Required for Herpes Simplex Virus Reactivation. Cell Host Microbe 2015, 18, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Du, T.; Zhou, G.; Roizman, B. Induction of apoptosis accelerates reactivation of latent HSV-1 in ganglionic organ cultures and replication in cell cultures. Proc. Natl. Acad. Sci. USA 2012, 109, 14616–14621. [Google Scholar] [CrossRef] [Green Version]
- Alkharsah, K.R.; Dedicoat, M.; Blasczyk, R.; Newton, R.; Schulz, T.F. Influence of HLA alleles on shedding of Kaposi sarcoma-associated herpesvirus in saliva in an African population. J. Infect. Dis. 2007, 195, 809–816. [Google Scholar] [CrossRef]
- Alkharsah, K.R.; Alzahrani, A.J.; Obeid, O.E.; El-Harith, E.H.; Guella, A.; Mohamed, E.A.; Haykal, A.H.; Stuhrmann, M.; Al-Ali, A.K. Vascular endothelial growth factor A polymorphism and risk of Kaposi’s sarcoma herpesvirus viremia in kidney allograft recipients. Transpl. Infect. Dis. 2014, 16, 783–789. [Google Scholar] [CrossRef]
- Houldcroft, C.J.; Kellam, P. Host genetics of Epstein–Barr virus infection, latency and disease. Rev. Med. Virol. 2015, 25, 71–84. [Google Scholar] [CrossRef] [Green Version]
- Sezgin, E.; An, P.; Winkler, C.A. Host Genetics of Cytomegalovirus Pathogenesis. Front. Genet. 2019, 10, 616. [Google Scholar] [CrossRef] [Green Version]
- Moraru, M.; Cisneros, E.; Gómez-Lozano, N.; de Pablo, R.; Portero, F.; Cañizares, M.; Vaquero, M.; Roustán, G.; Millán, I.; López-Botet, M.; et al. Host genetic factors in susceptibility to herpes simplex type 1 virus infection: Contribution of polymorphic genes at the interface of innate and adaptive immunity. J. Immunol. 2012, 188, 4412–4420. [Google Scholar] [CrossRef] [Green Version]
- Kriesel, J.D.; Bhatia, A.; Thomas, A. Cold sore susceptibility gene-1 genotypes affect the expression of herpes labialis in unrelated human subjects. Hum. Genome Var. 2014, 1, 14024. [Google Scholar] [CrossRef] [Green Version]
- Crosslin, D.R.; Carrell, D.S.; Burt, A.; Kim, D.S.; Underwood, J.G.; Hanna, D.S.; Comstock, B.A.; Baldwin, E.; de Andrade, M.; Kullo, I.J.; et al. Genetic variation in the HLA region is associated with susceptibility to herpes zoster. Genes Immun. 2015, 16, 1–7. [Google Scholar] [CrossRef] [Green Version]

| Sub Family | Virus | Cell Tropism | Latent Reservoir | Major Latent Transcripts | Inducers of Lytic Cycle |
|---|---|---|---|---|---|
| Alphaherpesvirinae | HSV1 HSV2 |
neurons fibroblasts |
neurons | LATs: sRNA1 and 2, lncRNAs, miRNAs |
immunosuppression, stress, UV hypoxia local tissue trauma hypothermia and hyperthermia cytokines and prostaglandins * hormonal changes NGF withdrawal Dexamethasone, NaBu |
| VZV | neurons | neurons | ORF63 VLT |
immunosuppression, stress, UV NGF withdrawal PI3K inhibitor, NaBu |
|
| Betaherpesvirinae | HCMV | fibroblasts PBMCs macrophages dendritic cells endothelial cells |
CD34+ HSCs CD33+ myeloid progenitors CD14+ monocytes NSCs |
UL138, UL111A LUNA US28 IE1x4 lncRNA2.7 and 4.9 UL144 |
immunosuppression proinflammatory cytokines * differentiation of cells allogeneic T-cell activation TPA |
| HHV-6 HHV-7 |
T cells | CD34+ HSCs PBMCs |
HHV-6: H6LTs:ORF99, ORF142, ORF145 U94 miR-U86 HHV-7: no data available |
immunosuppression infection with other pathogen * (HHV-6B) T-cell activation (HHV-7) TPA (HHV-6A/6B, HHV-7), TSA (HHV-6A) |
|
| Gammaherpesvirinae | EBV | B cells epithelial cells |
B cells | Latency I: EBNA1 Latency II: EBNA1 and LMP1 and/or 2 Latency III: EBNA1, 2, 3a, 3c, 3b, LP and LMP1, 2a and 2b |
immunosuppression, stress, chemotherapy, radiotherapy, hypoxia infection with other pathogen * prostaglandins * BCR crosslinking, TPA, NaBu, TSA |
| KSHV | B cells endothelial cells |
B cells | LANA (ORF73) vCyc (ORF72) vFLIP (ORF71) Kaposins vIRF3 (PEL and MCD) miRNAs |
immunosuppression, UV proinflammatory cytokines * hypoxia infection with other pathogen * TPA, NaBu, TSA, VPA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weidner-Glunde, M.; Kruminis-Kaszkiel, E.; Savanagouder, M. Herpesviral Latency—Common Themes. Pathogens 2020, 9, 125. https://doi.org/10.3390/pathogens9020125
Weidner-Glunde M, Kruminis-Kaszkiel E, Savanagouder M. Herpesviral Latency—Common Themes. Pathogens. 2020; 9(2):125. https://doi.org/10.3390/pathogens9020125
Chicago/Turabian StyleWeidner-Glunde, Magdalena, Ewa Kruminis-Kaszkiel, and Mamata Savanagouder. 2020. "Herpesviral Latency—Common Themes" Pathogens 9, no. 2: 125. https://doi.org/10.3390/pathogens9020125