-
PDF
- Split View
-
Views
-
Cite
Cite
Thomas C. Greenough, Gregory J. Babcock, Anjeanette Roberts, Hector J. Hernandez, William D. Thomas, Jennifer A. Coccia, Robert F. Graziano, Mohan Srinivasan, Israel Lowy, Robert W Finberg, Kanta Subbarao, Leatrice Vogel, Mohan Somasundaran, Katherine Luzuriaga, John L. Sullivan, Donna M. Ambrosino, Development and Characterization of a Severe Acute Respiratory Syndrome—Associated Coronavirus—Neutralizing Human Monoclonal Antibody That Provides Effective Immunoprophylaxis in Mice, The Journal of Infectious Diseases, Volume 191, Issue 4, 15 February 2005, Pages 507–514, https://doi.org/10.1086/427242
Close - Share Icon Share
Abstract
Background. Severe acute respiratory syndrome (SARS) remains a significant public health concern after the epidemic in 2003. Human monoclonal antibodies (MAbs) that neutralize SARS-associated coronavirus (SARSCoV) could provide protection for exposed individuals.
Methods. Transgenic mice with human immunoglobulin genes were immunized with the recombinant major surface (S) glycoprotein ectodomain of SARS-CoV. Epitopes of 2 neutralizing MAbs derived from these mice were mapped and evaluated in a murine model of SARS-CoV infection.
Results. Both MAbs bound to S glycoprotein expressed on transfected cells but differed in their ability to block binding of S glycoprotein to Vero E6 cells. Immunoprecipitation analysis revealed 2 antibody-binding epitopes: one MAb (201) bound within the receptor-binding domain at aa 490–510, and the other MAb (68) bound externally to the domain at aa 130–150. Mice that received 40 mg/kg of either MAb prior to challenge with SARS-CoV were completely protected from virus replication in the lungs, and doses as low as 1.6 mg/kg offered significant protection.
Conclusions. Two neutralizing epitopes were defined for MAbs to SARS-CoV S glycoprotein. Antibodies to both epitopes protected mice against SARS-CoV challenge. Clinical trials are planned to test MAb 201, a fully human MAb specific for the epitope within the receptor-binding region.
Severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) emerged as a major public health concern in 2003 [1, 2]. Most recently, 9 cases were confirmed in China, after 2 laboratory workers became infected [3]. Containing the outbreak required the quarantine of >1000 individuals who had been exposed to case patients, which created major disruption and concerns. This recent experience highlights not only the effectiveness of infection-control measures but also the difficulties in the detection of primary case patients and the tracking of contacts. With each new outbreak, the potential for worldwide dissemination remains.
Vaccines are in development and have shown effectiveness in animal models [4–6]. In the absence of an outbreak, however, the administration of a vaccine to the general population is unlikely. Therefore, the development of strategies to prevent infection and/or disease in unvaccinated, at-risk populations is crucial. Even a strategy that modifies the course of the disease and reduces virus burden without preventing infection may have a major impact on public health.
Neutralizing antibodies have proved to be effective in preventing viral infection in humans and are used as prophylaxis against varicella, hepatitis A, hepatitis B, rabies, and respiratory syncytial virus (RSV) infection [7]. Coronaviruses have a major surface (S) spike glycoprotein that mediates binding and entry of the virus into host cells [8–10]. The S glycoprotein interacts with a specific cellular receptor and, consequently, defines the host range and cytotropism of coronaviruses [11]. Angiotensin-converting enzyme 2 (ACE2), a metallopeptidase, has been identified as a SARS-CoV receptor, and its pattern of tissue expression matches the sites of virus recovery from infected individuals [12]. We, as well as others [13, 14], have shown that the minimal region of binding of the SARS-CoV S glycoprotein is contained within aa 270–510. We therefore have targeted this region in the development of neutralizing antibodies for use in the prevention of SARS and possibly the treatment of patients with SARS. In the present study, we describe the characterization of monoclonal antibodies (MAbs) directed against 2 epitopes on the S glycoprotein of SARS-CoV and their efficacy in protecting mice against challenge with live virus.
Materials and Methods
Codon-optimized S glycoprotein expression and purification. The amino acid sequence of the SARS-CoV S glycoprotein (Urbani strain, National Center for Biotechnology Information [strain no. AAP13441]) was used to design a codon-optimized version of the gene encoding the ectodomain of the S glycoprotein (aa 1–1190 [S1190]), as described elsewhere [13]. The synthetic gene was cloned into pcDNA3.1 Myc/His (Invitrogen) in frame with c-Myc (human proto-oncogene) and 6-histidine (His) epitope tags that enabled detection and purification. A similar approach was used to synthesize a codon-optimized gene encoding full-length S glycoprotein (S1255). Truncated soluble S glycoproteins were generated by polymerase chain reaction (PCR) amplification of the desired fragments from the vector encoding S1190. The cloned genes were sequenced to confirm that no errors had accumulated during the PCR process.
All constructs were transfected into human epithelial kidney (HEK)-293T/17 cells by use of lipofectamine 2000 (Invitrogen). Filtered supernatants from transfected cells were mixed with nickel-nitrilotriacetic acid (Ni-NTA) agarose (Invitrogen), and column filtration and protein elution using 250 mmol/L imidazole were done.
Mouse immunization and isolation of hybridomas. HuMAb mice (Medarex) are transgenic for human immunoglobulin genes, and mouse heavy-chain immunoglobulin genes are inactivated. HuMAb mice were injected weekly with 10 mg S1190, by use of complete Freund's as the primary adjuvant followed by incomplete Freund's, for a total of 6–8 weeks. ELISA was used to measure serum responses to S1190, and animals were killed when serum responses reached a plateau. Hybridomas were generated by fusion of splenocytes and partner cells, at a ratio of 6:1. Hybridoma supernatants were screened for reactivity to S1190, by ELISA, and proteins were purified by use of protein A sepharose beads (Amersham).
S1255 expression and staining with human MAbs. The construct encoding the entire codon-optimized SARS-CoV S glycoprotein (S1255) was transfected into HEK-293T/17 cells by use of lipofectamine 2000. Transfectants were harvested 48 h after transfection and were incubated with various concentrations of MAbs. Binding was detected by use of secondary-labeled goat anti-human IgG and flow cytometry using FACScan with CellQuest software (Becton Dickinson).
Detection of binding of S glycoprotein to Vero E6 cells. Vero E6 cells were resuspended in PBS containing 5% fetal calf serum and a c-Myc epitope-tagged protein consisting of the first 590 aa of the SARS-CoV S glycoprotein (S590) at a concentration of 10 nmol/L. Reactions were incubated for 1 h at room temperature in the presence or absence of varying concentrations of the MAbs. Binding was detected by use of anti- c-Myc antibody (9E10; BD Biosciences Pharmingen), followed by phycoerythrin-labeled goat anti-mouse IgG (Jackson), and was analyzed by means of flow cytometry using FACScan with CellQuest software.
Immunoprecipitation and Western blot. Constructs encoding soluble secreted S130, S150, S170, S269, S470, S490, S510, S270–510, or S1190 (described in figure 3) were transfected into HEK-293T/17 cells by use of lipofectamine 2000. Supernatant was harvested, filtered, and incubated with purified MAb and protein A sepharose beads. Precipitated proteins were resolved by SDS-PAGE and transferred to Imobilon P (Millipore), and membrane-bound protein was detected by use of 0.1 μg/mL anti-His (C-term) antibody (Invitrogen), followed by a 1:5000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Jackson), treatment with enhanced chemiluminescence (ECL) reagent (Amersham), and autoradiography.
Epitope mapping by immunoprecipitation. Truncated fragments of surface (S) glycoprotein of severe acute respiratory syndrome (SARS)—associated coronavirus (right) were tagged with 6-histidine (His) epitopes and were immunoprecipitated with monoclonal antibody (MAb) 68 (top left), MAb 201 (middle left), or nickel-nitrilotriacetic acid (Ni-NTA) agarose (bottom left); resolved by SDS-PAGE; transferred to solid support; and then visualized using anti-His (C-term) antibody, followed by a 1:5000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG, treatment with enhanced chemiluminescence reagent, and autoradiography. In the right panel, epitopes are depicted for MAb 201 (490–510 [blackened boxes]) and MAb 68 (130–150 [shaded boxes]).
S glycoprotein fragments from supernatants were precipitated with Ni-NTA agarose, resolved by SDS-PAGE, and transferred to Imobilon P for Western blotting. Membrane-bound protein was detected by use of 1 μg/mL MAb, followed by anti-human IgG HRP, and by ECL reagent and autoradiography.
Neutralizing-antibody assays. The neutralizing activity of MAbs was measured by use of an assay adapted from the work of Witte et al., as cited in the Office International des Epizooties Manual of Diagnostic Tests and Vaccines for Terrestrial Animals [15, 16]. Vero E6 cells were seeded at 5000 cells/well, in 96-well microtiter plates, on assay day -1 in a volume of 100 μL. On assay day 0, antibody dilutions were preincubated for 1 h with 100 TCID50 of virus stock (Urbani strain; generously provided by Larry Anderson [Centers for Disease Control and Prevention, Atlanta]). These mixtures of virus and antibody dilutions then were added to cells in replicates of 2 or 3. One additional set of antibody dilutions without virus was included as a control, to detect toxicity. Positive and negative controls (rabbit anti-S1190 and rabbit preimmune serum, respectively) were included in each assay. Virus stock was back titrated in each assay, to ensure that the inoculum was 30–300 TCID50/well. Presence or absence of cytopathic effect (CPE) after 72 h of incubation was determined by microscopy. Neutralizingantibody assays to titer mouse serum were done in a microneutralization assay using 2-fold dilutions of heat-inactivated serum. Serum samples were tested for the presence of antibodies that neutralized the infectivity of 100 TCID50 of SARSCoV in Vero E6 cell monolayers, by use of 4 wells per dilution on a 96-well plate. The presence of viral CPE was read on days 3 and 4. The dilution of serum that completely prevented CPE in 50% of the wells was calculated by means of the Reed- Muench formula [17]. After microscopic visualization of CPE, medium was replaced by PBS, CellTiter96 reagent (Promega) was added, and plates were incubated for 2–4 h until gradations of color between uninfected and infected controls were easily distinguished visually. CellTiter96 is metabolized to a soluble, colored product, the concentration of which is proportional to the number of viable cells in the culture. Absorbance is reduced in wells with significant CPE. To inactivate virus, 10% SDS was added, and the absorbance (optical density measured at 490 nm) was read by use of a universal plate reader (EL 800; Bio-Tek Instruments). Percent protective effect was calculated as follows: 100(observed - maximum CPE)/(minimum CPE x maximum CPE), where “maximum CPE” refers to absorbance in control wells with virus and no MAb and “minimum CPE” refers to absorbance in control wells with no virus and no MAb.
Protection of mice with MAbs. The mouse studies were approved by the Animal Care and Use Committee of the National Institutes of Health (Bethesda, MD) and were done in an approved animal biosafety level 3 facility. The experiments were done as described elsewhere [18]. In brief, 4–6-week-old female BALB/c mice (Taconic) were housed in cages with 4 mice per cage. Mice received 400 μL MAb or serum by intraperitoneal (ip) injection. The mice were bled the next day to determine the level of neutralizing antibody achieved, and lightly anesthetized mice were challenged with 105 TCID50 of SARS-CoV, administered intranasally. The mice were killed 2 days later, and lung and nasal turbinate tissues were homogenized in a 10% or 5% (wt/vol) suspension (lung or nasal turbinate, respectively) in Leibovitz 15 medium (Invitrogen) complemented with l-glutamine (Gibco), piperacillin (Sigma Aldrich), gentamicin (Invitrogen), and amphotericin B (Quality Biological) at final concentrations of 4 mmol/L, 0.4 mg/L, 0.1 mg/L, and 5 mg/L, respectively.Virus titers (expressed as TCID50 per gram) were determined on Vero E6 cell monolayers in 24- or 96-well plates. The lower limits of detection in lung and nasal turbinate tissues were 101.5 and 101.8 TCID50/g, respectively.
Biacore assay. Surface plasmon resonance technology [19] was used to determine the affinity of MAbs to S590, a fragment that includes the entire receptor-binding domain. In brief, each MAb at a concentration of 5 μg/mL was captured on the surface of a Biacore chip that had been coated with goat anti-human Fc polyclonal antibody. The captured MAbs were exposed to various concentrations of S590 in solution that flowed through the chip, and association and dissociation rates were measured by the changes in concentration of biomolecules at the interface of the chip and the media. Association ([kon = 1/Ms] x 104, where M indicates moles per liter and s indicates seconds) and dissociation ([koff = 1/s] x 10-4) rate constants were calculated; from these, the dissociation constant KD ([koff/kon = M] x 10-9) was derived [19, 20].
Statistics. Log-transformed virus titers were compared in a nonparametric, 2-tailed Mann-Whitney test, and P > .05 was considered to be statistically significant.
Results
Generation and characterization of SARS-CoV-neutralizing MAbs. To generate neutralizing human MAbs directed against SARS-CoV, transgenic mice with active human immunoglobulin genes (HuMAb mice) were immunized with the soluble ectodomain of SARS-CoV S glycoprotein (S1190; as described elsewhere [13]). From 36 hybridomas that produced S1190-specific antibodies, 2 MAbs (201 and 68) with potent neutralizing activity were purified and fully characterized. Peptide sequence analysis (by Edman degradation) and light-chain analysis (by ELISA) of MAb 201 demonstrated a fully human IgG1 antibody with a single κ light chain. MAb 68 was a chimeric molecule with a human heavy chain and murine light chain (λ).
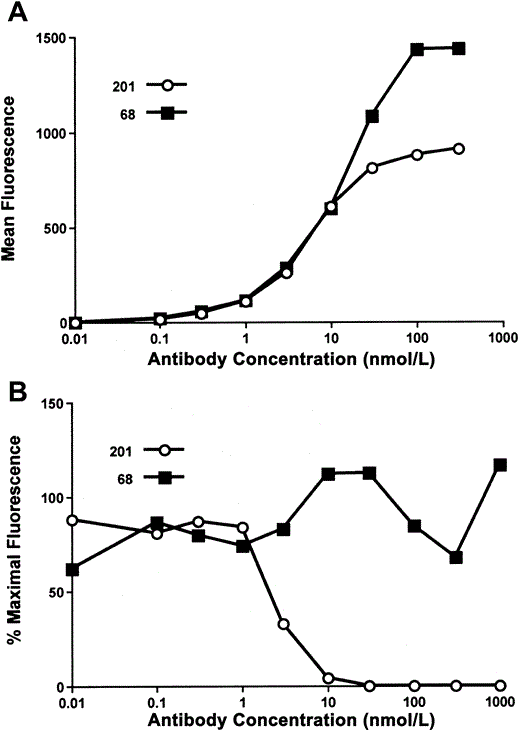
To ensure that the MAbs selected were reactive to full-length, membrane-bound S glycoprotein, we synthesized a codon-optimized gene encoding full-length SARS-CoV S glycoprotein (S1255), for expression on HEK-293T/17 cells. The dose-dependent binding curves for both MAbs suggested similar avidity, with 50% maximal binding at 1 nmol/L (figure 1A). MAb 201 bound S1255 with a lower maximal intensity than did MAb 68, suggesting differences in protein recognition. In a flow cytometry-based assay, MAb 201 specifically blocked binding of S590 (aa 1–590) to Vero E6 cells, whereas MAb 68 showed no blocking activity (figure 1B). Affinity measurements (Biacore assay) obtained by use of soluble S glycoprotein fragment S590 and immobilized MAb showed affinity constants (KD) of 34 nmol/L and 83 nmol/L for MAbs 201 and 68, respectively.
In vitro characterization of monoclonal antibodies (MAbs) 201 and 68. A, Results of flow-cytometry analysis of human epithelial kidney—293T/17 cells expressing full-length surface (S) glycoprotein (S1255) of severe acute respiratory syndrome (SARS)-associated coronavirus. Cells were stained with either MAb 201 (circles) or MAb 68 (squares), followed by phycoerythrin-labeled goat anti-human IgG. B, Results for Vero E6 cells incubated with 10 nmol/L of a c-Myc (human proto-oncogene) epitope-tagged protein consisting of the first 590 aa of S glycoprotein (S590), in the presence of various concentrations of MAb 201 (circles) or MAb 68 (squares). S glycoprotein binding was detected via the epitope tag using the murine anti-c-Myc antibody 9E10, followed by phycoerythrin-labeled goat anti-mouse IgG. Reduction in fluorescent intensity mediated by the MAb was calculated and plotted.
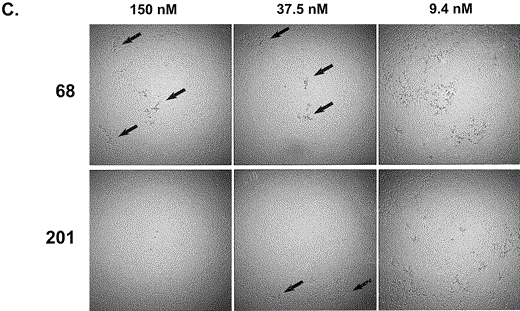
Characterization of in vitro neutralization of SARS-CoV by MAbs 201 and 68. To measure the neutralizing potency of each MAb, we adapted microtiter assays used by diagnostic laboratories to measure neutralizing antibodies to various animal coronaviruses [15]. The determination of neutralizing activity included a colorimetric readout of metabolically active cells (CellTiter96), as well as the visually observed level of CPE in the microtiter wells. Both MAbs provided a 50% maximal protective effect (colorimetric assay) at concentrations of ~ 1 nmol/L, in repeated assays (representative assay shown in figure 2A). Although neutralization, as measured by cell metabolism, clearly showed protective effects with both MAbs, microscopic inspection of the monolayers showed subtle differences that suggested infection of cells even in the presence of the highest concentrations of MAb 68. Specifically, tiny foci of CPE were observed in nearly every well with a high concentration of MAb 68. In contrast, no CPE was observed in the wells with high concentrations of MAb 201 (figure 2B). In microneutralization assays using a 30-fold higher-concentration virus inoculum (3000 TCID50/well), the cytoprotective effects of the 2 MAbs were again indistinguishable (colorimetric assay), although curves were shifted slightly toward higher MAb concentrations (data not shown). With the higher-concentration virus inoculum, the small foci of CPE seen in the presence of the highest concentration of MAb 68 were more numerous than with the standard-concentration virus inoculum (100 TCID50/well); however, complete protection from CPE was again observed with the highest concentration of MAb 201 (figure 2C).
Patterns of inhibition of cytopathic effects (CPEs) in microneutralization assays. A, Results of infection of Vero E6 cells with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV), preincubated with either monoclonal antibody (MAb) 201 (circles) or MAb 68 (squares) at various concentrations. After 3 days of incubation, CellTiter96 reagent was added to measure the metabolic activity of cells. Protective effect was calculated as described in Materials and Methods and plotted. B, Dilution series for each MAb incubated with SARS-CoV and subsequently added to Vero E6 cells in triplicate. Three days postinfection, individual wells were graded by the percentage of cells involved in the CPE, as shown in the key (right). C, Photographs demonstrating results of microscopic examination of infected Vero E6 cells. This experiment was identical to that described for panel B, with the exception that the virus inoculum was 3000 TCID50/well, instead of the standard 100 TCID50/well. Arrows indicate small foci of CPE observed in the presence of MAb 68 at 150 nmol/L but not of MAb 201 at 150 nmol/L. More-extensive CPE was seen in the presence of both MAbs at the lowest concentration of 9.4 nmol/L.
Determination of binding epitopes for MAbs 201 and 68. To map the epitopes recognized by the MAbs, we synthesized genes encoding S glycoproteins truncated at the carboxy- and aminoterminal domains (figure 3, right). Two distinct patterns of immunoprecipitation were observed: (1) MAb 68 (figure 3, top left) precipitated all fragments, including S150, except S130 and S270–510, demonstrating recognition of an epitope within aa 130–150; and (2) MAb 201 (figure 3, middle left) precipitated fragments S510, S270–510, and S1190 but no N-terminal fragment with <510 aa, demonstrating recognition of an epitope within aa 490–510. Both MAbs displayed identical recognition patterns on Western blots of denatured S glycoprotein fragments, demonstrating that linear epitopes were recognized (data not shown).
Immunoprophylactic efficacy of MAbs 201 and 68 in a mouse model of virus challenge. To evaluate the protective efficacy of these MAbs in vivo, we used an established murine model of SARS-CoV infection. This model has been used to demonstrate protection by neutralizing antibodies that develop in mice previously challenged with live SARS-CoV [18] or after vaccination with inoculum that includes expressed S glycoprotein [4, 6]. Mice were given purified MAb, by ip injection, on the day before challenge with 105 TCID50 of SARS-CoV (Urbani strain) and were killed 2 days after challenge. Nonimmune sera from uninfected mice and an irrelevant MAb (Palivizumab [Medimmune], which is specific for RSV infection) were used as controls. Blood was collected just prior to challenge, and serum neutralizing titers in mice treated with 40 mg/kg MAb 201 were determined to be 1:16. The neutralization of virus by sera from mice treated with MAb 68 was incomplete; at the lowest dilutions, CPE was noted to be limited and patchy, which was similar to the pattern of neutralization by MAb 68 described in figure 2.
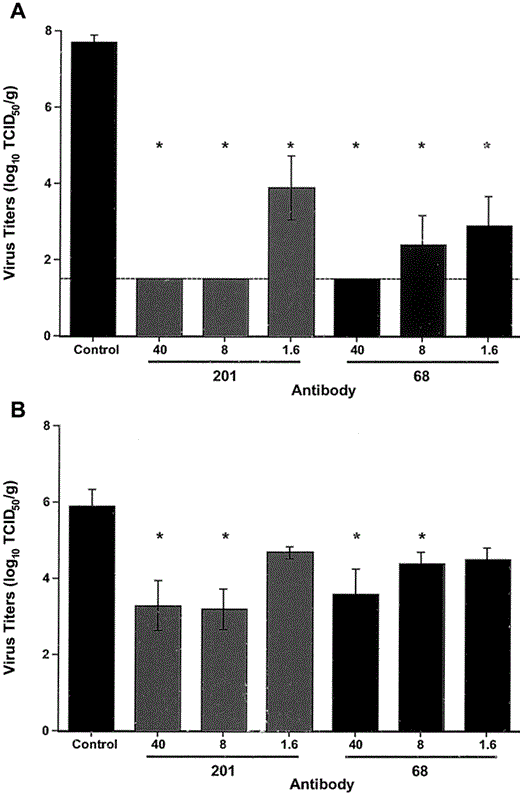
Virus titers were measured in lung and nasal turbinate tissues 48 h after challenge. At doses of ~1.6–40 mg/kg, both MAbs provided significant protection from replication of virus in lung tissues (P = .02; figure 4A). Virus was undetectable in lung tissues from mice treated with 40 or 8 mg/kg MAb 201 (>106-fold reduction in virus load). All groups of mice treated with 40 or 8 mg/kg of either MAb had significantly lower titers of virus in nasal turbinate tissues (upper respiratory tract), compared with control mice treated with irrelevant MAb (P ⩽ .03), but the reduction in virus replication was less profound than that observed in lung tissues (figure 4B). A suggestion of a greater protective effect with MAb 201 than with MAb 68 was found, as judged by the differences in the reduction of virus titers at 8 mg/kg; however, the number of observations was too small to show significant differences between the MAbs.
Efficacy of immunoprophylaxis in mice challenged with severe acute respiratory syndrome (SARS)—associated coronavirus (SARS-CoV). Mice (4 in each group) were treated with the indicated monoclonal antibody (MAb), at estimated doses of 40, 8, and 1.6 mg/kg, 1 day prior to intranasal challenge with 105 TCID50 of SARS-CoV. Two days after infection, lung tissue (A) or nasal turbinate tissue (B) was harvested and homogenized, and in vitro virus-titration assays were performed. Virus titers are shown as mean log10 TCID50 per gram of tissue, with standard errors. Comparisons of the results between groups were made by use of the nonparametric, 2-tailed Mann-Whitney test. *P > .05, compared with control. The limit of detection of the assay is indicated (A, dashed line).
Discussion
Here we describe the characteristics of 2 MAbs that effectively neutralize SARS-CoV in vitro and recognize 2 distinct linear epitopes on the SARS-CoV S glycoprotein, 1 within and 1 external to theminimal receptor-binding domain. Despite these differences in protein recognition, both MAbs protected mice in a model of virus challenge. The mechanism of neutralization for MAb 201, which bound to aa 490–510, is most likely a direct interference of binding of virus S glycoprotein to its receptor, ACE2. Neutralization and protection of mice by MAb 68, which bound to aa 130–150 but failed to block S590 binding to Vero E6 cells, must involve a different mechanism. The simplest explanation would be interference with the conformational changes that are required for virus-cell membrane fusion to occur after binding to ACE2. Additional possible mechanisms include (1) alterations in physical properties of SARSCoV (e.g., aggregation) and (2) interference with an as-yetunrecognized coreceptor.
The immunoprophylactic efficacies of these MAbs in the murine model were defined by significant reductions in virus load in lung and nasal turbinate tissues. The large inoculum of SARS-CoV in these experiments was used to ensure infection, so that these effects could be best defined.We emphasize that this model does not allow conclusions regarding modification of SARS or prevention of transmission of SARS-CoV. Nonetheless, a reasonable hypothesis is that these effects could be achieved, given the results of preclinical and clinical testing of the MAb Palivizumab in the prevention of RSV disease. In animal models of RSV infection, Palivizumab does not prevent infection but significantly reduces virus titers [20]. Large clinical trials have established that modification of disease and prevention of transmission can be achieved by treating infants at risk for RSV infection and disease with Palivizumab (reviewed in [21]).
The protection of mice by MAbs 201 and 68 shows a dose dependence, with a >2-log reduction (>99%) of SARS-CoV in lung tissue even at the lowest dose (1.6 mg/kg). Palivizumab as a preventive measure against RSV infection provides a benchmark to gauge the feasibility of achieving effective doses of SARS-CoV-neutralizing MAb in humans. At doses of 15 mg/kg, Palivizumab provides effective prophylaxis against RSV infection in humans. Results of our animal studies suggest that the MAbs that neutralize SARS-CoV could be used at dose levels sufficient to significantly reduce virus titers in humans.
Having demonstrated effective protection by these MAbs against SARS-CoV challenge in the murine model, extension of these studies to include additional animal models with demonstrated pathology will be important. Future nonhuman primate studies also will examine the therapeutic efficacy of these and other MAbs, after infection has been established [22]. Early administration of a neutralizing MAb might be therapeutically effective, since peaks in virus replication in infected humans occur 10–14 days after onset of symptoms, prior to the appearance of substantial neutralizing-antibody titers. Given the lag between symptom onset and maximum virus concentrations in lungs, there may be an opportunity to identify and treat individuals with neutralizing MAbs.
We have selected the human MAb 201 that targets the receptor-binding domain of S glycoprotein for initial clinical studies of safety and pharmacokinetics. The S glycoprotein epitope recognized by this MAb has not demonstrated any sequence variability in epidemiological studies and, therefore, may be a stable target for neutralization in vivo [23–25].
The use of antibodies to prevent a variety of viral infections has proved to be a successful strategy [7]. The rapidity of development and the safety record of human MAbs suggest a primary role for this modality in response to SARS and other future emerging infectious diseases.
Acknowledgements
We express special thanks to Larry Anderson (Centers for Disease Control and Prevention, Atlanta), for supplying virus, viral lysate, and serum samples from convalescing patients. We also acknowledge Daniel Carraher, James Coderre, Katherine Donahue, Diana Esshaki, and Heather Walker, for excellent technical assistance, and Frank Brewster, for lab management and advice.




![Epitope mapping by immunoprecipitation. Truncated fragments of surface (S) glycoprotein of severe acute respiratory syndrome (SARS)—associated coronavirus (right) were tagged with 6-histidine (His) epitopes and were immunoprecipitated with monoclonal antibody (MAb) 68 (top left), MAb 201 (middle left), or nickel-nitrilotriacetic acid (Ni-NTA) agarose (bottom left); resolved by SDS-PAGE; transferred to solid support; and then visualized using anti-His (C-term) antibody, followed by a 1:5000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG, treatment with enhanced chemiluminescence reagent, and autoradiography. In the right panel, epitopes are depicted for MAb 201 (490–510 [blackened boxes]) and MAb 68 (130–150 [shaded boxes]).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/jid/191/4/10.1086/427242/2/m_191-4-507-fig003.gif?Expires=1716439100&Signature=ca9dUDHNiZVaCkwGC9osS18kqhLK03ta5IbyIi6jIwqDTrbyq34Ts2w1NTBR2U~IOWbmOkQIIJcMOcqpTktOwU0yx8nSLSYMLacEWUY5aSObQ7kicUzZF8MeykguXDndP3E-Wk-sSXo8q2yHgRKRIP2vpDhX6p1sh4QUcGTFmd6iI9T~lAFrxM44PR2zkgUwBbbd0~NXUGJ3NrddD9ayc9z0zT1FrjS26naJibTH4ig0oM1n-4U43QJLq-Oyd9I2H62f~pWMM1JYmeCFlHYYJU2RmsBLe0NAr4iVvjAU4EuI6MeCzROwMgHgMKpqFNq3s4Uopub5fsg~ULZ0zMuPDw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)