Abstract
A practical colorimetric assay was developed for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). For this purpose, magnetic γ Fe2O3 nanoparticles were synthesized and used as a peroxidase-like mimic activity molecule. In the presence of γ Fe2O3 nanoparticles, the color change of H2O2 included 3,3′,5,5′-tetramethylbenzidine was monitored at the wavelength of 654 nm when spike protein interacted with angiotensin-converting enzyme 2 receptor. This oxidation-reduction reaction was examined both spectroscopically and by using electrochemical techniques. The experimental parameters were optimized and the analytical characteristics investigated. The developed assay was applied to real SARS-CoV-2 samples, and very good results that were in accordance with the real time polymerase chain reaction were obtained.
Graphical abstract

Similar content being viewed by others
Introduction
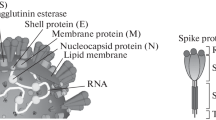
The latest coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or COVID-19 is the seventh member of the coronavirus family [1]. Like the other previous two viruses in this family, namely, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 is a positive-sense, non-segmented, and an enveloped RNA virus [2]. It has single strands of RNA and crown-like spikes on the outer surface [3]. Four types of main proteins, namely, spike (S), membrane, envelope, and nucleocapsid [4], are the proteins that are found in SARS-CoV-2. The glycoprotein, S-protein of SARS-CoV-2, is made up of 2 subunits, namely, S1 and S2. S1 has the receptor-binding domain (RBD) part which provides the attachment of the virus to the receptor while S2 facilitates the virus cell membrane fusion [3, 5,6,7]. S1 subunit has a kind of open conformation which helps it to interact with angiotensin-converting enzyme 2 (ACE2) [7,8,9]. ACE2 is found in the membranes of the lungs, arteries, heart, kidney, and intestine cells [10, 11]. ACE2 also acts as an entry point for some coronaviruses, such as SARS-CoV and SARS-CoV-2 [12].
Although SARS-CoV and MERS have higher fatality rates compared to SARS-CoV-2 [13], since SARS-CoV-2 is more infectious, the overall number of deaths is much higher than SARS-CoV and MERS.
For this reason, early and effective detection of SARS-CoV-2 becomes more important day by day. Currently, laborious, time-consuming, and specialized personnel needed real-time polymerase chain reaction (RT-PCR) technique is the gold standard method for SARS-CoV-2 detection. Due to the disadvantages mentioned above, accurate, rapid, and practical methods that have a point of care (POC) nature are urgently needed [14]. Conducting the diagnostic test at home without needing to leave the house and learning the result rapidly, will allow a person to quarantine himself immediately thus preventing the spread of the virus. Apart from these, it is also stated that there are asymptomatic or paucysymptomatic cases in the early stages of the infection, and such cases are the cause of transmission [15,16,17]. In this context, for SARS-CoV-2 detection, many attempts have been made in terms of appropriate biosensor developments, which are usually based on antigen-antibody interactions [4, 14, 18]. Also, suitable rapid tests have been developed and used for this purpose [19]. The highly insufficient sensitivity value in rapid antibody tests, which was seen as a hope in active case detection, also decreases the value of these tests [19].
Herein, we manage to develop a novel colorimetric assay for SARS-CoV-2 detection. The interactions are based on the infection mechanism of SARS-CoV-2 which we believe differs from the other antigen-based rapid tests or biosensors. For this purpose, we used magnetic γ Fe2O3 nanoparticles as a peroxidase-like mimic activity providing molecule [20] and monitor 3,3′,5,5′-tetramethylbenzidine (TMB) colorimetric reagent oxidation-reduction reactions [21, 22] when S-protein interacted with ACE2 receptor. Lastly, developed assay was applied for real sample analysis where the validation was done via RT-PCR method.
Experimental
Materials and instruments
All reagents were of analytical grade and used without any purification. Sodium hydroxide (NaOH), hydrogen chloride (HCl), nitric acid (HNO3), and potassium dihydrogen phosphate (KH2PO4) were purchased from Merck (www.merck.millipore.com). Iron(III)chloride hexahydrate (FeCl3.6H2O), iron dichloride tetrahydrate (FeCl2.4H2O), and TMB liquid substrate (Cat. No. T0440) were purchased from Sigma-Aldrich (www.sigmaaldrich.com). Recombinant human ACE2 was obtained from RayBiotech (www.raybiotech.com). S-protein was purchased from Biovision (www.biovision.com). Influenza viruses were obtained from HyTest (www.hytestfi.com). All solutions were prepared with double deionized ultrapure water.
T60UV modeled PG instruments limited UV-Vis spectrophotometer was used to measure the absorbance of samples by using 1 cm quartz cuvettes. Scanning electron microscopy (SEM) image and energy dispersive X-ray spectroscopy (EDS) measurements were obtained from JSM-7600 F FEG-SEM at 15.0 kV. X-ray Powder Diffraction (XRD) measurements were carried out with the Rigaku Smartlab XRD apparatus.
For RT-PCR, “Bio-Speedy® SARS-CoV-2 + VOC202012/01 RT-qPCR” kit was used which was obtained from Bioeksen R&D Technologies Istanbul-Turkey. The amplification was performed on a CFX96 Touch Real-Time PCR Detection System Instrument (Bio-Rad Laboratories, Inc., USA). vNAT® tubes were also used from Bioeksen R&D Technologies Istanbul-Turkey (extraction consumables of vNAT® Viral Nucleic Acid Buffer Cat No BS-NA-510; vNAT® Transfer Tube Cat No BS-NA-513-100-Bioeksen R&D Technologies Istanbul-Turkey).
The enzyme-mimicking activity of γ Fe2O3
Synthesis and characterization experiments of γ Fe2O3 magnetic nanoparticles are detailed in the supplementary file [23, 24].
The peroxidase-like mimic activity of magnetic γ Fe2O3 nanoparticle in the presence of hydrogen peroxide (H2O2) was well known [20, 25,26,27]. Herein, we used TMB (Cat. No. T0440) that has already contained H2O2, a ready to use Enzyme-Linked Immuno Sorbent Assay (ELISA) test [28, 29]. For this reason, in our assay, the oxidation of colorless TMB solution to the blue color oxidized TMB(Ox) solution was obtained without adding any H2O2 to the medium. For clear observation of color change, 1 mL of TMB solution and 50 μL of magnetic γ Fe2O3 nanoparticle colloid were mixed and waited for 1 h. During this process, the solution was constantly in contact with air, and intermittent shaking was carried out to prevent the magnetic γ Fe2O3 nanoparticles from settling to the bottom. In the presence of magnetic γ Fe2O3 nanoparticles, the oxidation progress of TMB to TMB(Ox) was monitored by measuring the absorbance of the reaction mixture using a UV-visible spectrophotometer in a range of 450–800 nm.
Colorimetric detection assay of S-protein
For determination of S-protein, the first blue-colored TMB(Ox) form was obtained by mixing 1 mL of TMB (optimized amount, data not shown) and 50 μL (10 mg.mL−1) of magnetic γ Fe2O3 nanoparticles [24] in the Eppendorf tube for 1 h. After TMB was completely transformed into TMB(Ox), 100 μL from 10 ng.mL−1 of ACE2 receptor (the optimal binding function concentration specified in the data sheet) was added into the blue colored mixture. Then, different concentrations of S-protein solution were added to the blue colored mixture and incubated constantly in contact with air and intermittent shaking for 30 min. For UV measurements, at the end of the incubation time, a colorless or light blue colored mixture was transferred to a 1 cm path length quartz cuvette, and a magnet was placed under the cuvette to keep the magnetic γ Fe2O3 nanoparticles at the bottom of the cuvette. Magnetic separation was performed before each UV-vis measurement to avoid scattering caused by magnetic nanoparticles (Scheme 1). For the real sample analysis, after completing ACE2 incubation, real samples were added to the medium instead of S-protein.
The tentative mechanism of the developed sensing assay for colorimetric determination of SARS-CoV-2 S-protein with TMB colorimetric reagent. a A structure of SARS-CoV-2. b Oxidation procedure of TMB (H2O2 included) in the presence of magnetic γ Fe2O3 nanoparticles. c Working principle of developed sensing assay
Selectivity studies
Selectivity studies of the developed colorimetric SARS-CoV-2 biosensor were performed with influenza A virus. H3N2 Brisbane virus and H1N1 New Caledonia virus were selected as model influenza A viruses. In order to observe the influenza viruses interfering effects, 48.70 ng.mL−1 virus solutions and S-protein solution were added to the solution that contained 1 mL of TMB, 50 μL of magnetic γ Fe2O3 nanoparticles, and 100 μL ACE2 (from 10 ng.mL−1 ACE2 solution). After waiting at 25 °C for 30 min, experiments were carried out in triplicate.
Real sample application
Study design
Collection of specimens
Combined oropharyngeal and nasopharyngeal swab specimens were collected from samples received at the microbiology department of the Muğla Training and Research Hospital, a tertiary hospital of more than 600 beds in Muğla. The samples included in the study were selected from those who were requested to be tested in the routine COVID-19 diagnostic procedure in the laboratory.
RT-PCR
The validity of our colorimetric biosensor was tested by analyzing real samples that were previously investigated by RT-PCR. The threshold level was chosen as 200 RFU (relative fluorescence units) to calculate the number of threshold cycle (Cq) in accordance with the manufacturer’s recommendations. The samples were collected from 41 positive SARS-CoV-2 patients for which Cq ≤ 33 and 41 negative patients (no Cq detection). Combined nasopharyngeal and throat samples in vNAT® tubes were admitted to the laboratory for SARS Cov-2 PCR tests of 41 patients with positive PCR test results and 41 healthcare personnel who were SARS Cov-2 PCR test negative in routine institution screening. Samples were stored at 2–8 °C approximately for 48 h until they were tested. All samples were run on the same day of experiment.
Interpretation of colorimetric test results
The samples were evaluated with the naked eye according to the degree of color change in the test wells (Fig. 5B). The scoring system (that was used to evaluate the test results) dependence to the level of color change is shown in Table 1.
Determination of sample size
Statistical power analysis was made by using the “G. Power-3.1.9.4” program. The Chi-square Goodness-of Fit Test was applied using the study rates of Ventura et al., and the standardized effect size (0.3667410) was calculated [30]. It has been calculated that a minimum of 82 people will be recruited to this study with type 1 error (alpha error) probability of 5% and 80% power at a moderate effect size and 1 degree of freedom.
Results and discussion
The importance of practical and effective assay for SARS-CoV-2 detection is undeniable nowadays. For the fabrication of our approach, first, we synthesized and characterized γ Fe2O3 magnetic nanoparticles (Fig. S1). Then, we formed our diagnostic assay step by step as described in the experimental part (Scheme 1).
The tentative mechanism of the interaction
It is well known that iron oxides (γ Fe2O3 and Fe3O4) mimic the peroxidase and catalyze activity in the presence of H2O2 [20, 25,26,27]. Herein, we prefer to use H2O2 included TMB solution to simplify the procedure. γ Fe2O3 acts like a peroxidase enzyme, and TMB is oxidized in the presence of H2O2 (Scheme 1). In order to investigate this situation further, cyclic voltammograms for each step were monitored as presented in Fig. 1A. As it can be seen from Fig. 1A, a, TMB has two well-defined oxidation (0.41 V) and reduction peaks (0.34 V). On the other hand, for γ Fe2O3, an oxidation at 0 V and reduction peaks at more negative potential than −0.4 V are seen (Fig. 1A, b). When these two were mixed in the presence of H2O2, the oxidation of TMB happens causing a blue color and more significant anodic and cathodic peaks (Fig. 1A, c). ACE2 introduction to the above mixture causes a little bit decrease at the peaks (Fig. 1A, d) while after the binding of S-protein, drastic decrease especially at the oxidation peak but also at the reduction peak is observed (Fig. 1A, d).
Cyclic voltammograms and UV spectra with the color difference photos of the developed system for each step. A a 1 mL of TMB; b 5 μL of γ Fe2O3; c 1 mL TMB + 50 μL γ Fe2O3; d 1 mL TMB + 50 μL γ Fe2O3/100 μL from 10 ng.mL−1 of ACE2 receptor; e 1 mL TMB + 50 μL γ Fe2O3/100 μL from 10 ng.mL−1 of ACE2 receptor/95.20 ng.mL−1 of S-protein at a carbon paste electrode in 10 mL of phosphate buffer, pH = 7.4, scan rate: 0.10 V s − 1. B a 1 mL of TMB; b 50 μL of magnetic γ Fe2O3 nanoparticle solution (no UV spectra because of scattering); c 1 mL of TMB/50 μL of magnetic γ Fe2O3 nanoparticles; d 1 mL of TMB/50 μL of magnetic γ Fe2O3 nanoparticles/100 μL from 10 ng.mL−1 of ACE2; e 1 mL of TMB/50 μL of magnetic γ Fe2O3 nanoparticles/100 μL from 10 ng.mL−1 of ACE2/100 μL of 100 mmol.L−1 pH:7.4 phosphate buffer; f 1 mL of TMB/50 μL of magnetic γ Fe2O3 nanoparticles/95.20 ng.mL−1 of S-protein; g 1 mL of TMB/50 μL of magnetic γ Fe2O3 nanoparticles/100 μL from 10 ng.mL−1 of ACE2/95.20 ng.mL−1 of S-protein
Besides these, we examined the UV absorbance spectra of our colorimetric assay as demonstrated in Fig. 1B. Figure 1B a, belongs to the TMB spectrum with almost zero absorbance. The UV spectrum of γ Fe2O3 could not be obtained due to the scatterings (not shown in the UV spectra). On the other hand, the highest absorbance was obtained for TMB(ox) corresponding to blue color at the wavelength of 654 nm when γ Fe2O3 and TMB were mixed in the presence of H2O2 (Fig. 1B, c). The addition of ACE2 and S-protein separately did not change the absorbance too much (Fig. 1B, d and f) while the simultaneous addition of these two molecules in the same medium decreased the absorbance significantly (Fig. 1B, g).
Based on the previous findings in the literature, it has been known that there is a thiol−disulfide balance on the cell surface [31]. On the other hand, when the RBD part of the S-protein binds to the ACE2 receptor, the cysteine pair of S-protein switches between disulfide classes upon binding of the RBD to the receptor. Also, the two cysteine pairs are located parallelly and closely together and may involve in disulfide shuffling [9]. We believe that TMB here acts like an oxidoreductase molecule and causes a reduction of disulfide bonds to thiols upon binding of RBD to the receptor. In another study conducted by Xue et al., it is demonstrated that the addition of cysteine in TMB (ox) solution causes the reduction of the molecule [32]. At this point, we can not explain the exact reason for color disappearance when the binding occurs, but it might be due to the favorable alignment of cysteine molecules as mentioned above. This may also explain the reason of occurance of no color disappearance when only S-protein or ACE2 presented in the medium.
Optimization of detection parameters
For obtaining the best results, the optimizations of reaction temperature and incubation time have been performed and given in Fig. 2. All optimization experiments were carried out by using 1 mL of TMB, 50 μL of γ Fe2O3, 100 μL from 10 ng.mL−1 of ACE2 receptor, and 24.60 ng.mL−1 of S-protein in the Eppendorf tubes. It is important to have a POC system that works at 25 °C considering its nature. From Fig. 2a, it is clear that at 37 °C, we get better results. However, at 25 °C, it is still possible to see the color difference with the naked eye. For practicality, we used 25 °C as our applied temperature and used this value for further studies.
Meanwhile for the incubation time, apart from not being the best value, since the drastic changes occurred at 30 min and considering fast response of developed assay, we took 30 min as our optimized incubation time and used it for further studies (Fig. 2b).
Analytical characteristics
Under the optimized working conditions, we examined the analytical characteristics of our assay. As can be seen from Fig. 3a, a decrease in the absorbance at the wavelength of 654 nm of blue color occurs in proportion with the increase in the concentration of S-protein. By using the decreasing absorbance values obtained due to the increase in S-protein concentration, the linear response range was determined between 4.98 ng.mL−1 and 139.00 ng.mL−1 with an equation of y = −0.0115x + 1.8978, R2:0.99 (Fig. 3b). Limit of detection (LOD) value was determined by taking the lowest concentration value in the calibration graph which is 4.98 ng.mL−1 [33]. Lastly, relative standard deviation (R.S.D) value was estimated as %3.16 ± 0.04 for 24.60 ng.mL−1 (n: 3 times). On the other hand, analytical range of the method including the UV spectrum of blank solution is given in Fig. 3c.
Analytical characteristics experiments were performed under the optimized working conditions. a UV spectra and color difference photos, according to increasing concentrations of S-protein from 4.89 ng.mL−1 to 139.00 ng.mL−1. b Linear range. c Analytical range of the developed colorimetric sensor at the wavelength of 654 nm
Selectivity studies
As shown in Fig. 4A, when S-protein and influenza A viruses were mixed at the same concentrations, although a loss in the blue color was observed for S-protein (Fig. 4, d), there was no significant color change for solutions containing influenza A viruses (Fig. 4, b and c). This result is expected since the Influenza virus is not a virus that interacts with the ACE2 receptor. Also, this results demonstrate that our diagnostic system is capable of discriminating COVID-19 from influenza viruses.
A UV spectra and color difference photos of a 1 mL TMB + 50 μL γ Fe2O3 + 100 μL ACE2 from 10 ng.mL−1 of ACE2 receptor solution, b 1 mL TMB + 50 μL γ Fe2O3 + 100 μL from 10 ng.mL−1 of ACE2 receptor solution +48.70 ng.mL−1 H1N1 virus, c 1 mL TMB + 50 μL γ Fe2O3 + 100 μL from 10 ng.mL−1 of ACE2 receptor solution +48.70 ng.mL−1 H3N2 virus, d 1 mL TMB + 50 μL γ Fe2O3 + 100 μL from 10 ng.mL−1 of ACE2 receptor solution +48.70 ng.mL−1 S-protein. B Bar chart graphs showing the differences in absorbance values obtained with influenza viruses and S-protein. All the selectivity experiments were performed at 25°C for 30 min
Real sample application
Determination of sample size
Since RT-PCR was accepted as the gold standard for this assessment, positive and negative samples by molecular techniques were accepted as true positive and true negative samples, successively (Table 2).
Overall, amongst 41 positive samples, our colorimetric assay detected 40 samples as definitive positive (Fig. 5A). Amongst the 41 negative samples with RT-PCR test, our colorimetric biosensor detected 37 samples as definitive negative. Actually, when the results of the gold standard were compared with the colorimetric method, positive reaction was observed in 40 of 41 PCR positive patients, while the test result of one PCR positive sample was recorded as borderline. On the other hand, borderline results were observed in 2 of 41 PCR negative samples, and positive test results were found in 2 of 41 PCR negative samples with the colorimetric method. While calculating the performance properties of our test, these borderline results, that were determined with developed colorimetric biosensor, were accepted as false-negative results for the PCR-positive sample group and also were accepted as false-positive results for the PCR-negative sample group.
Picture of A the positive and negative sample colorimetric analysis results. B Scoring system of developed colorimetric assay. All the real sample experiments were carried out by adding 100 μL fresh sample to 100 μL of test solution including 1 mL of TMB, 50 μL of magnetic γ Fe2O3 nanoparticles, and 100 μL from 10 ng.mL−1 ng of ACE2 at 25 °C waiting for 30 min
Meanwhile, alpha variant (in the lineage B.1.1.7), a variant of SARS-CoV-2 that was first seen in Great Britain, was detected in 34 of 41 RT-PCR positive samples. As it is well known, because of huge number of mutations that are usually found in its S-protein, this variant is more transmissible compared to the other previously observed variants [34].
Overall, a similar positive reaction was seen for the results that were obtained with colorimetric biosensor independent from the presence of alpha variant. For this reason, it is obvious that the mutations that the virus has undergone over time do not have any bad influence on our colorimetric responses.
Based on the parameters at Table 2, the performance characteristics including sensitivity, accuracy, positive and negative predictive values, positive and negative likelihood ratios (LRs), and selectivity of our colorimetric biosensors were calculated and demonstrated in Table 3.
As it is well known, as LR values differ much more from the value of 1.0, the possibility of their corporation with the presence or absence of disease increases. Because of this, tests with very high LR+ and very low LR− can be very discerning, and as a result, tests with LRs >10 or < 0.1 helps us to constitute or to eliminate a diagnosis [35].
Based on the receiver operating characteristic (ROC) analysis, area under curve (AUC) has been estimated as 0.939 (p value = 0.000 < 0.05) while the confidence interval of the area under curve is found as 95%CI = [0.88, 0.99].
Overall, the ability to distinguish the test is presented by the area under the ROC curve. According to the ROC analysis results that we obtained, it can be said that the discrimination of our rapid colorimetric test method is very good. Also for real samples, the color change began at 10th min and was completed at 20th min on the contrary of 30 min that had been seen in biosensor development procedure.
Conclusion
The need for effective, fast, and practical detection of SARS-CoV-2 is very important nowadays. Herein, we manage to develop a practical approach for SARS-CoV-2 diagnosis. The ease and practicality of the method make it an important candidate for becoming an effective diagnostic kits for SARS-CoV-2 detection in future. On the other hand, considering the mutations that happen until now, our detection assay will be beneficial because there is no need to change antigens or any other reagents like primer reagents, etc. The application of developed colorimetric assay to the real samples resulted with very good agreement with PCR method. Also, the successful detection of alpha variant of COVID-19 demonstrated that our assay is capable to detect the variants because of its interaction mechanism. Considering the performance characteristics of the developed colorimetric assay, it can be concluded that the system is a serious candidate to be a rapid screening test in the diagnosis of suspected cases of COVID-19.
References
Antiochia R (2020) Nanobiosensors as new diagnostic tools for SARS, MERS and COVID-19: from past to perspectives. Microchim Acta 187:1–13. https://doi.org/10.1007/s00604-020-04615-x
Farzin L, Sadjadi S, Sheini A, Mohagheghpour E (2021) A nanoscale genosensor for early detection of COVID-19 by voltammetric determination of RNA-dependent RNA polymerase (RdRP) sequence of SARS-CoV-2 virus. Microchim Acta 188:1–12. https://doi.org/10.1007/s00604-021-04773-6
Astuti I, Ysrafil (2020) Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr Clin Res Rev 14:407–412. https://doi.org/10.1016/j.dsx.2020.04.020
Eissa S, Alhadrami HA, Al-Mozaini M et al (2021) Voltammetric-based immunosensor for the detection of SARS-CoV-2 nucleocapsid antigen. Microchim Acta 188:199. https://doi.org/10.1007/s00604-021-04867-1
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- a n update on the status. Mil Med Res 7:11. https://doi.org/10.1186/s40779-020-00240-0
Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. In: Coronaviruses: Methods and Protocols. Springer, New York, pp 1–23
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. https://doi.org/10.1016/j.cell.2020.02.058
Song W, Gui M, Wang X, Xiang Y (2018) Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog 14:e1007236. https://doi.org/10.1371/journal.ppat.1007236
Meirson T, Bomze D, Markel G (2021) Structural basis of SARS-CoV-2 spike protein induced by ACE2. Bioinformatics 37:929–936. https://doi.org/10.1093/bioinformatics/btaa744
Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203:631–637. https://doi.org/10.1002/path.1570
Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, Breitbart RE, Acton S (2000) A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res 87:E1–E9. https://doi.org/10.1161/01.res.87.5.e1
ACE2 angiotensin converting enzyme 2 [Homo sapiens (human)] - Gene - NCBI. https://www.ncbi.nlm.nih.gov/gene/59272. Accessed 25 Jun 2021
Layqah LA, Eissa S (2019) An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim Acta 186:1–10. https://doi.org/10.1007/s00604-019-3345-5
Rahmati Z, Roushani M, Hosseini H, Choobin H (2021) Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Microchim Acta 188:1–9. https://doi.org/10.1007/s00604-021-04762-9
Bordi L, Nicastri E, Scorzolini L et al (2020) Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS-CoV-2), Italy, February 2020. Eurosurveillance 25:2000170
Chan JFW, Yip CCY, To KKW, et al (2020) Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol 58: e00310. https://doi.org/10.1128/JCM.00310-20
Wei WE, Li Z, Chiew CJ, et al (2020) Presymptomatic Transmission of SARS-CoV-2 — Singapore, January 23–March 16, 2020. MMWR 69:411. https://doi.org/10.15585/mmwr.mm6914e1
Kim HY, Lee JH, Kim MJ, Park SC, Choi M, Lee W, Ku KB, Kim BT, Changkyun Park E, Kim HG, Kim SI (2021) Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens Bioelectron 175:112868. https://doi.org/10.1016/j.bios.2020.112868
Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H (2020) Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol 129:104455. https://doi.org/10.1016/j.jcv.2020.104455
Nasir M, Nawaz MH, Latif U, Yaqub M, Hayat A, Rahim A (2017) An overview on enzyme-mimicking nanomaterials for use in electrochemical and optical assays. Microchim Acta 184:323–342
Ding Y, Zhao J, Li B, Zhao X, Wang C, Guo M, Lin Y (2018) The CoOOH-TMB oxidative system for use in colorimetric and test strip based determination of ascorbic acid. Microchim Acta 185:1–10. https://doi.org/10.1007/s00604-018-2675-z
Li F, Hu Y, Zhao A, Xi Y, Li Z, He J (2020) β-Cyclodextrin coated porous Pd@Au nanostructures with enhanced peroxidase-like activity for colorimetric and paper-based determination of glucose. Microchim Acta 187:1–11. https://doi.org/10.1007/s00604-020-04410-8
Lin Y, Xu G, Wei F, Zhang A, Yang J, Hu Q (2016) Detection of CEA in human serum using surface-enhanced Raman spectroscopy coupled with antibody-modified Au and γ-Fe2O3@Au nanoparticles. J Pharm Biomed Anal 121:135–140. https://doi.org/10.1016/j.jpba.2016.01.027
Kalyoncu D, Buyuksunetci YT, Anık Ü (2019) Development of a sandwich immunosensor for concurrent detection of carcinoembryonic antigen (CEA), vascular endothelial growth factor (VEGF) and Α-fetoprotein (AFP) biomarkers. Mater Sci Eng C 101:88–91. https://doi.org/10.1016/j.msec.2019.03.079
Mitra K, Ghosh AB, Sarkar A, Saha N, Dutta AK (2014) Colorimetric estimation of human glucose level using γ-Fe 2O3 nanoparticles: an easily recoverable effective mimic peroxidase. Biochem Biophys Res Commun 451:30–35. https://doi.org/10.1016/j.bbrc.2014.07.028
Roy A, Sahoo R, Ray C, Dutta S, Pal T (2016) Soft template induced phase selective synthesis of Fe2O3 nanomagnets: one step towards peroxidase-mimic activity allowing colorimetric sensing of thioglycolic acid. RSC Adv 6:32308–32318. https://doi.org/10.1039/c6ra00963h
Wu S, Guo D, Xu X, Pan J, Niu X (2020) Colorimetric quantification and discrimination of phenolic pollutants based on peroxidase-like Fe3O4 nanoparticles. Sensors Actuators B Chem 303:127225. https://doi.org/10.1016/j.snb.2019.127225
Jouve N, Despoix N, Espeli M, Gauthier L, Cypowyj S, Fallague K, Schiff C, Dignat-George F, Vély F, Leroyer AS (2013) The involvement of CD146 and its novel ligand galectin-1 in apoptotic regulation of endothelial cells. J Biol Chem 288:2571–2579. https://doi.org/10.1074/jbc.M112.418848
Harpaz D, Eltzov E, Ng TSE, Marks RS, Tok AIY (2020) Enhanced colorimetric signal for accurate signal detection in paper-based biosensors. Diagnostics 10:28. https://doi.org/10.3390/diagnostics10010028
Della VB, Cennamo M, Minopoli A et al (2020) Colorimetric test for fast detection of SARS-COV-2 in nasal and throat swabs. ACS Sensors 5:3043–3048. https://doi.org/10.1021/acssensors.0c01742
Hati S, Bhattacharyya S (2020) Impact of thiol-disulfide balance on the binding of COVID-19 spike protein with angiotensin-converting enzyme 2 receptor. ACS Omega 5:16292–16298. https://doi.org/10.1021/acsomega.0c02125
Xue Z, Xiong L, Rao H, Liu X, Lu X (2019) A naked-eye liquid-phase colorimetric assay of simultaneous detect cysteine and lysine. Dyes Pigments 160:151–158. https://doi.org/10.1016/j.dyepig.2018.07.054
Wang K, Fan D, Liu Y, Wang E (2015) Highly sensitive and specific colorimetric detection of cancer cells via dual-aptamer target binding strategy. Biosens Bioelectron 73:1–6. https://doi.org/10.1016/j.bios.2015.05.044
Center DC (2021) SARS-CoV-2 Variant Classifications and Definitions. In: Cdc. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html. Accessed 25 Jun 2021
Ranganathan P, Aggarwal R (2018) Understanding the properties of diagnostic tests – Part 2: Likelihood ratios. Perspect Clin Res 9:99. https://doi.org/10.4103/picr.picr_41_18
Funding
This work was supported by BAP Project department of Mugla Sıtkı Kocman University with the project number of 20/108/02/1/4. Also for this study, the patent application has been made with the application number of 21/00352.
Author information
Authors and Affiliations
Contributions
Yudum Tepeli Büyüksünetçi and Ülkü Anık “contributed” to the development and optimization of colorimetric biosensor together with analytical characteristics examinations. They are also involved at the real sample analysis with newly developed biosensor. Burak Ekrem Çitil and Utku Tapan were responsible from the real sample collections and analysis of them with the newly developed biosensor system and PCR methods.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 151 kb)
Rights and permissions
About this article
Cite this article
Büyüksünetçi, Y.T., Çitil, B.E., Tapan, U. et al. Development and application of a SARS-CoV-2 colorimetric biosensor based on the peroxidase-mimic activity of γ-Fe2O3 nanoparticles. Microchim Acta 188, 335 (2021). https://doi.org/10.1007/s00604-021-04989-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04989-6