Flavones’ and Flavonols’ Antiradical Structure–Activity Relationship—A Quantum Chemical Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Conformer Geometry Generation
2.2. Radical Geometry Generation
2.3. Quantitative Determinants of Antioxidant Potential
2.3.1. Hydrogen Atom Transfer Mechanism
2.3.2. Electron Transfer–Proton Transfer Mechanism
2.3.3. Sequential Proton Loss–Electron Transfer Mechanism
2.3.4. Mechanism-Independent Determinants
3. Results and Discussion
3.1. Molecule and Radical Electronic Structure Investigation
3.2. Morin
3.3. Mechanisms of Action in Terms of the Determinants
3.3.1. Hydrogen Atom Transfer Mechanism
3.3.2. Electron Transfer–Proton Transfer Mechanism
3.3.3. Sequential Proton Loss–Electron Transfer Mechanism
3.3.4. Mechanism-Independent Determinants
3.3.5. Antioxidant Capacity Summary
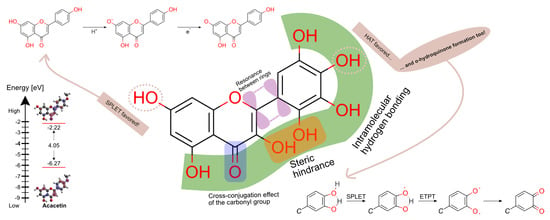
3.4. Intramolecular Hydrogen Bonding
3.5. Catechol Moiety
3.5.1. Intramolecular Hydrogen Swap
-
ΔrG0(298K) is Gibb’s free energy of the reaction, at 298 K (25 °C) and pressure of 1 atmosphere.
-
ε0 is total electronic energy [Hartree].
-
Gcorr is thermal free energy [Hartree].
3.5.2. Diradical Formation
3.5.3. o-Hydroquinone Formation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Electron Density Swap

Appendix B. Frontier Molecular Orbitals Theory
Appendix B.1. Highest Occupied (HOMO) and Lowest Unoccupied (LUMO) Molecular Orbitals
| Flavonoids | EHOMO | ELUMO | |η| | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | ||||
| Flavones | Acacetin | −6.270 (M) | −2.217 | 4.572 | 4.265 | |||||
| Apigenin | −6.319 (L) | −2.220 | 4.263 | 4.512 | 4.255 | |||||
| Chrysin | −6.460 (L) | −2.320 | 4.456 | 4.170 | ||||||
| Chrysoeriol | −6.135 (M) | −2.221 | 4.374 | 4.625 | 4.319 | |||||
| Diosmetin | −6.151 (M) | −2.222 | 4.439 | 4.624 | 4.310 | |||||
| Genkwanin | −6.329 (L) | −2.195 | 4.245 | 4.565 | ||||||
| Luteolin | −6.215 (M) | −2.240 | 4.393 | 4.213 | 4.565 | 4.297 | ||||
| Flavonols | Fisetin | −5.924 (H) | −2.250 | 4.413 | 4.487 | 4.581 | 4.398 | |||
| Galangin | −6.172 (M) | −2.373 | 4.394 | 4.514 | 4.288 | |||||
| Kaempferol | −5.982 (H) | −2.283 | 4.415 | 4.491 | 4.631 | 4.374 | ||||
| Morin | −6.141 (M) | −2.285 | 4.505 | 4.350 | 4.270 | 4.534 | 4.295 | |||
| Myricetin | −5.941 (H) | −2.315 | 4.527 | 4.377 | 4.399 | 4.503 | 4.638 | 4.396 | ||
| Quercetin | −5.934 (H) | −2.304 | 4.544 | 4.340 | 4.518 | 4.647 | 4.400 | |||
Appendix B.2. Single Occupied Molecular Orbital (SOMO)
| Flavonoids | ESOMO | |||||||
|---|---|---|---|---|---|---|---|---|
| C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | ||
| Flavones | Acacetin | −6.367 | −6.502 | |||||
| Apigenin | −6.570 | −6.456 | −6.586 | |||||
| Chrysin | −6.699 | −6.925 | ||||||
| Chrysoeriol | −6.470 | −6.205 | −6.320 | |||||
| Diosmetin | −6.471 | −6.216 | −6.319 | |||||
| Genkwanin | −6.594 | −6.429 | ||||||
| Luteolin | −6.494 | −6.557 | −6.343 | −6.440 | ||||
| Flavonols | Fisetin | −6.329 | −6.290 | −6.212 | −6.195 | |||
| Galangin | −6.463 | −6.429 | −6.573 | |||||
| Kaempferol | −6.356 | −6.230 | −6.176 | −6.336 | ||||
| Morin | −6.237 | −6.489 | −6.400 | −6.305 | −6.445 | |||
| Myricetin | −6.202 | −6.246 | −6.265 | −6.268 | −6.102 | −6.258 | ||
| Quercetin | −6.189 | −6.224 | −6.292 | −6.122 | −6.270 | |||
| Luteolin | Fisetin | Myricetin | Quercetin | |
|---|---|---|---|---|
| C3’-C4’ | C3’-C4’ | C3’-C4’ | C4’-C5’ | C4’-C5’ |
 −6.704 |
 −5.989 |
 −6.049 |
 −6.003 |
 −6.032 |
 −6.893 |
 −7.184 |
 −6.721 |
 −6.757 |
 −6.866 |
References
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal 2012, 24, 981–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamanaka, R.B.; Chandel, N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 2010, 35, 505–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, L.; Franco, M.C.; Estevez, A.G. Reactive nitrogen species in cellular signaling. Exp. Biol. Med. 2015, 240, 711–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, H.M. An essential role for free radicals and derived species in signal transduction. FASEB J. 1997, 11, 118–124. [Google Scholar] [CrossRef]
- Breen, A.P.; Murphy, J.A. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 1995, 18, 1033–1077. [Google Scholar] [CrossRef]
- Pratt, D.A.; Tallman, K.A.; Porter, N.A. Free radical oxidation of polyunsaturated lipids: New mechanistic insights and the development of peroxyl radical clocks. Acc. Chem. Res. 2011, 44, 458–467. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Gebicki, J.M. Proteins are major initial cell targets of hydroxyl free radicals. Int. J. Biochem. Cell Biol. 2004, 36, 2334–2343. [Google Scholar] [CrossRef]
- Hulsmans, M.; Van Dooren, E.; Holvoet, P. Mitochondrial reactive oxygen species and risk of atherosclerosis. Curr. Atheroscler. Rep. 2012, 14, 264–276. [Google Scholar] [CrossRef]
- Vera-Ramirez, L.; Sanchez-Rovira, P.; Ramirez-Tortosa, M.C.; Ramirez-Tortosa, C.L.; Granados-Principal, S.; Lorente, J.A.; Quiles, J.L. Free radicals in breast carcinogenesis, breast cancer progression and cancer stem cells. Biological bases to develop oxidative-based therapies. Crit. Rev. Oncol. Hematol. 2011, 80, 347–368. [Google Scholar] [CrossRef]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Johnson, I.T.; Saltmarsh, M. Polyphenols: Antioxidants and beyond. Am. J. Clin. Nutr. 2005, 81, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.A.; Bolling, B.W. A review of the efficacy of dietary polyphenols in experimental models of inflammatory bowel diseases. Food Funct. 2015, 6, 1773–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Teng, H.; Xie, Z.; Cao, H.; Cheang, W.S.; Skalicka-Woniak, K.; Georgiev, M.I.; Xiao, J. Modifications of dietary flavonoids towards improved bioactivity: An update on structure–activity relationship. Crit. Rev. Food Sci. Nutr. 2018, 58, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Masek, A.; Chrzescijanska, E.; Latos, M.; Zaborski, M. Influence of hydroxyl substitution on flavanone antioxidants properties. Food Chem. 2017, 215, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Musialik, M.; Kuzmicz, R.; Pawlowski, T.S.; Litwinienko, G. Acidity of hydroxyl groups: An overlooked influence on antiradical properties of flavonoids. J. Org. Chem. 2009, 74, 2699–2709. [Google Scholar] [CrossRef]
- Heijnen, C.G.M.; Haenen, G.R.M.M.; Van Acker, F.A.A.; Van Der Vijgh, W.J.F.; Bast, A. Flavonoids as peroxynitrite scavengers: The role of the hydroxyl groups. Toxicol. Vitr. 2001, 15, 3–6. [Google Scholar] [CrossRef]
- Amorati, R.; Valgimigli, L. Modulation of the antioxidant activity of phenols by non-covalent interactions. Org. Biomol. Chem. 2012, 10, 4147–4158. [Google Scholar] [CrossRef]
- Lucarini, M.; Pedulli, G.F.; Guerra, M. A Critical Evaluation of the Factors Determining the Effect of Intramolecular Hydrogen Bonding on the O-H Bond Dissociation Enthalpy of Catechol and of Flavonoid Antioxidants. Chem. A Eur. J. 2004, 10, 933–939. [Google Scholar] [CrossRef]
- Cano, A.; Williamson, G.; Garcia-Conesa, M.T. Superoxide scavenging by polyphenols: Effect of conjugation and dimerization. Redox Rep. 2002, 7, 379–383. [Google Scholar] [CrossRef]
- Milenković, D.; Dimitrić Marković, J.M.; Dimić, D.; Jeremić, S.; Amić, D.; Pirković, M.S.; Marković, Z.S. Structural characterization of kaempferol: A spectroscopic and computational study. Maced. J. Chem. Chem. Eng. 2019, 38, 49–62. [Google Scholar] [CrossRef]
- Lucarini, M.; Pedrielli, P.; Pedulli, G.F.; Cabiddu, S.; Fattuoni, C. Bond dissociation energies of O-H bonds in substituted phenols from equilibration studies. J. Org. Chem. 1996, 61, 9259–9263. [Google Scholar] [CrossRef]
- Yokozawa, T.; Chen, C.P.; Dong, E.; Tanaka, T.; Nonaka, G.I.; Nishioka, I. Study on the inhibitory effect of tannins and flavonoids against the 1,1-diphenyl-2-picrylhydrazyl radical. Biochem. Pharmacol. 1998, 56, 213–222. [Google Scholar] [CrossRef]
- Lin, C.; Zhu, C.; Hu, M.; Wu, A.; Zerendawa, B.; Suolangqimei, K. Structure-activity Relationships of Antioxidant Activity in vitro about Flavonoids Isolated from Pyrethrum Tatsienense. J. Intercult. Ethnopharmacol. 2014, 3, 123. [Google Scholar] [CrossRef]
- Amić, A.; Marković, Z.; Dimitrić Marković, J.M.; Stepanić, V.; Lučić, B.; Amić, D. Towards an improved prediction of the free radical scavenging potency of flavonoids: The significance of double PCET mechanisms. Food Chem. 2014, 152, 578–585. [Google Scholar] [CrossRef]
- Amić, A.; Marković, Z.; Klein, E.; Dimitrić Marković, J.M.; Milenković, D. Theoretical study of the thermodynamics of the mechanisms underlying antiradical activity of cinnamic acid derivatives. Food Chem. 2018, 246, 481–489. [Google Scholar] [CrossRef]
- Dimitrić Marković, J.M.; Pejin, B.; Milenković, D.; Amić, D.; Begović, N.; Mojović, M.; Marković, Z.S. Antiradical activity of delphinidin, pelargonidin and malvin towards hydroxyl and nitric oxide radicals: The energy requirements calculations as a prediction of the possible antiradical mechanisms. Food Chem. 2017, 218, 440–446. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Amić, D.; Davidović-Amić, D.; Bešlo, D.; Trinajstić, N. Structure-radical scavenging activity relationships of flavonoids. Croat. Chem. Acta 2003, 76, 55–61. [Google Scholar]
- Rasulev, B.F.; Abdullaev, N.D.; Syrov, V.N.; Leszczynski, J. A Quantitative Structure-Activity Relationship (QSAR) study of the antioxidant activity of flavonoids. QSAR Comb. Sci. 2005, 24, 1056–1065. [Google Scholar] [CrossRef]
- Van Acker, S.A.B.E.; Van Den Berg, D.J.; Tromp, M.N.J.L.; Griffioen, D.H.; Van Bennekom, W.P.; Van Der Vijgh, W.J.F.; Bast, A. Structural aspects of antioxidant activity of flavonoids. Free Radic. Biol. Med. 1996, 20, 331–342. [Google Scholar] [CrossRef]
- Bors, W.; Heller, W.; Michel, C.; Saran, M. Flavonoids as antioxidants: Determination of radical-scavenging efficiencies. Methods Enzymol. 1990, 186, 343–355. [Google Scholar] [PubMed]
- Galano, A.; Raúl Alvarez-Idaboy, J. Computational strategies for predicting free radical scavengers’ protection against oxidative stress: Where are we and what might follow? Int. J. Quantum Chem. 2019, 119, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Marković, Z. Study of the mechanisms of antioxidative action of different antioxidants. J. Serbian Soc. Comput. Mech. 2016, 10, 135–150. [Google Scholar] [CrossRef]
- Sroka, Z.; Żbikowska, B.; Hładyszowski, J. The antiradical activity of some selected flavones and flavonols. Experimental and quantum mechanical study. J. Mol. Model. 2015, 21, 307. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Allouche, A. Software News and Updates Gabedit—A Graphical User Interface for Computational Chemistry Softwares. J. Comput. Chem. 2012, 32, 174–182. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum mechanical continuum solvation models. Chem. Rev. 2005, 105, 2999–3093. [Google Scholar] [CrossRef]
- Mennucci, B.; Tomasi, J. Continuum solvation models: A new approach to the problem of solute’s charge distribution and cavity boundaries. J. Chem. Phys. 1997, 106, 5151–5158. [Google Scholar] [CrossRef]
- Aparicio, S. A systematic computational study on flavonoids. Int. J. Mol. Sci. 2010, 11, 2017–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todorova, T.Z.; Traykov, M.G.; Tadjer, A.V.; Velkov, Z.A. Structure of flavones and flavonols. Part I: Role of substituents on the planarity of the system. Comput. Theor. Chem. 2013, 1017, 85–90. [Google Scholar] [CrossRef]
- Wang, J.; Becke, A.D.; Smith, V.H. Evaluation of ⟨S2⟩ in restricted, unrestricted Hartree-Fock, and density functional based theories. J. Chem. Phys. 1995, 102, 3477–3480. [Google Scholar] [CrossRef]
- Tada, K.; Tanaka, S.; Kawakami, T.; Kitagawa, Y.; Okumura, M.; Yamaguchi, K. Spin contamination errors on spin-polarized density functional theory/plane-wave calculations for crystals of one-dimensional materials. Appl. Phys. Express 2019, 12, 115506. [Google Scholar] [CrossRef]
- Urbaniak, A.; Szelag, M.; Molski, M. Theoretical investigation of stereochemistry and solvent influence on antioxidant activity of ferulic acid. Comput. Theor. Chem. 2013, 1012, 33–40. [Google Scholar] [CrossRef]
- Glendening, E.D.; Reed, A.E.; Carpenter, J.E.; Weinhold, F. NBO.
- Limacher, P.A.; Lüthi, H.P. Cross-conjugation. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 477–486. [Google Scholar] [CrossRef]
- Jovanovic, S.V.; Steenken, S.; Hara, Y.; Simic, M.G. Which Ring in Flavonoids Is Responsible for Antioxidant Activity? J. Chem. Soc. Perkins Trans. 1996, 2, 2497–2504. [Google Scholar] [CrossRef]
- Hatzidimitriou, E.; Nenadis, N.; Tsimidou, M.Z. Changes in the catechin and epicatechin content of grape seeds on storage under different water activity (aw) conditions. Food Chem. 2007, 105, 1504–1511. [Google Scholar] [CrossRef]
- Çelik, S.E.; Özyürek, M.; Güçlü, K.; Apak, R. Solvent effects on the antioxidant capacity of lipophilic and hydrophilic antioxidants measured by CUPRAC, ABTS/persulphate and FRAP methods. Talanta 2010, 81, 1300–1309. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The Ferric Reducing Ability of Plasma (FRAP) as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, X.; Zhao, S.; Zhang, Y. Antioxidant-capacity-based models for the prediction of acrylamide reduction by flavonoids. Food Chem. 2015, 168, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Milenković, D.; Đorović, J.; Petrović, V.; Avdović, E.; Marković, Z. Hydrogen atom transfer versus proton coupled electron transfer mechanism of gallic acid with different peroxy radicals. React. Kinet. Mech. Catal. 2018, 123, 215–230. [Google Scholar] [CrossRef]
- Milenković, D.; Dorović, J.; Jeremić, S.; Dimitrić Marković, J.M.; Avdović, E.H.; Marković, Z. Free Radical Scavenging Potency of Dihydroxybenzoic Acids. J. Chem. 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Jeremić, S.; Radenković, S.; Filipović, M.; Antić, M.; Amić, A.; Marković, Z. Importance of hydrogen bonding and aromaticity indices in QSAR modeling of the antioxidative capacity of selected (poly)phenolic antioxidants. J. Mol. Graph. Model. 2017, 72, 240–245. [Google Scholar] [CrossRef]
- Filipović, M.; Marković, Z.; Dorović, J.; Marković, J.D.; Lučić, B.; Amić, D. QSAR of the free radical scavenging potency of selected hydroxybenzoic acids and simple phenolics. Comptes Rendus Chim. 2015, 18, 492–498. [Google Scholar] [CrossRef]
- Chen, Y.; Xiao, H.; Zheng, J.; Liang, G. Structure-thermodynamics-antioxidant activity relationships of selected natural phenolic acids and derivatives: An experimental and theoretical evaluation. PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef]
- Arora, A.; Nair, M.G.; Strasburg, G.M. Structure-activity relationships for antioxidant activities of a series of flavonoids in a liposomal system. Free Radic. Biol. Med. 1998, 24, 1355–1363. [Google Scholar] [CrossRef]
- Álvarez-Diduk, R.; Ramírez-Silva, M.T.; Galano, A.; Merkoçi, A. Deprotonation mechanism and acidity constants in aqueous solution of flavonols: A combined experimental and theoretical study. J. Phys. Chem. B 2013, 117, 12347–12359. [Google Scholar] [CrossRef] [PubMed]
- Dimitrić Marković, J.M.; Milenković, D.; Amić, D.; Mojović, M.; Pašti, I.; Marković, Z.S. The preferred radical scavenging mechanisms of fisetin and baicalein towards oxygen-centred radicals in polar protic and polar aprotic solvents. RSC Adv. 2014, 4, 32228–32236. [Google Scholar] [CrossRef]
- Huang, D.; Boxin, O.U.; Prior, R.L. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef] [PubMed]
- Dimitrić Marković, J.M.; Milenković, D.; Amić, D.; Popović-Bijelić, A.; Mojović, M.; Pašti, I.A.; Marković, Z.S. Energy requirements of the reactions of kaempferol and selected radical species in different media: Towards the prediction of the possible radical scavenging mechanisms. Struct. Chem. 2014, 25, 1795–1804. [Google Scholar] [CrossRef]
- Peng, C.; Bernhard Schlegel, H. Combining Synchronous Transit and Quasi-Newton Methods to Find Transition States. Isr. J. Chem. 1993, 33, 449–454. [Google Scholar] [CrossRef]
- Maini, S.; Hodgson, H.L.; Krol, E.S. The UVA and aqueous stability of flavonoids is dependent on B-ring substitution. J. Agric. Food Chem. 2012, 60, 6966–6976. [Google Scholar] [CrossRef]
- Pearson, R.G. Chemical hardness and density functional theory. J. Chem. Sci. 2005, 117, 369–377. [Google Scholar] [CrossRef]
- Parr, R.G.; Pearson, R.G. Absolute Hardness: Companion Parameter to Absolute Electronegativity. J. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
- Waki, T.; Nakanishi, I.; Matsumoto, K.I.; Kitajima, J.; Chikuma, T.; Kobayashi, S. Key role of chemical hardness to compare 2,2-diphenyl-1-picrylhydrazyl radical scavenging power of flavone and flavonol O-glycoside and C-glycoside derivatives. Chem. Pharm. Bull. 2012, 60, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Mazzone, G.; Russo, N.; Toscano, M. Antioxidant properties comparative study of natural hydroxycinnamic acids and structurally modified derivatives: Computational insights. Comput. Theor. Chem. 2016, 1077, 39–47. [Google Scholar] [CrossRef]





 |
||||||||
|---|---|---|---|---|---|---|---|---|
| Flavonoid | C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | |
| Flavones | Acacetin | -OCH3 | -OH | -OH | ||||
| Apigenin | -OH | -OH | -OH | |||||
| Chrysin | -OH | -OH | ||||||
| Chrysoeriol | -OCH3 | -OH | -OH | -OH | ||||
| Diosmetin | -OH | -OCH3 | -OH | -OH | ||||
| Genkwanin | -OH | -OH | -OCH3 | |||||
| Luteolin | -OH | -OH | -OH | -OH | ||||
| Flavonols | Fisetin | -OH | -OH | -OH | -OH | |||
| Galangin | -OH | -OH | -OH | |||||
| Kaempferol | -OH | -OH | -OH | -OH | ||||
| Morin | -OH | -OH | -OH | -OH | -OH | |||
| Myricetin | -OH | -OH | -OH | -OH | -OH | -OH | ||
| Quercitin | -OH | -OH | -OH | -OH | -OH | |||
| Flavonoid | θ C2–C1’ | θ● C2–C1’ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | |||
| Flavones | Acacetin | −13.7 | 12.9 (Δ = −2.2) |
7.9 (Δ = −7.2) |
|||||
| Apigenin | −15.5 | −5.7 (Δ = + 9.8) |
−12.5 (Δ = + 3.0) |
−8.6 (Δ = −6.9) |
|||||
| Chrysin | 20.6 | 18.7 (Δ = −1.9) |
16.3 (Δ = −4.3) |
||||||
| Chrysoeriol | −16.9 | −3.7 (Δ = + 13.2) |
−18.3 (Δ = −1.4) |
−4.9 (Δ = + 12.0) |
|||||
| Diosmetin | 16.7 | 12.1 (Δ = −4.6) |
17.6 (Δ = + 0.9) |
9.9 (Δ = −6.8) |
|||||
| Genkwanin | −16.0 | −4.5 (Δ = + 11.5) |
−14.7 (Δ = + 1.3) |
||||||
| Luteolin | 15.9 | 23.5 (Δ = + 7.6) |
12.2 (Δ = −3.7) |
19.2 (Δ = + 3.3) |
8.3 (Δ = −7.6) |
||||
| Flavonols | Fisetin | 8.8 | 3.3 (Δ = −5.5) |
0.0 (Δ = −8.8) |
0.3 (Δ = −8.5) |
6.2 (Δ = −2.6) |
|||
| Galangin | −15.0 | 0.0 (Δ = + 15.0) |
−3.6 (Δ = + 11.4) |
−14.2 (Δ = + 0.8) |
|||||
| Kaempferol | −3.5 | 0.0 (Δ = + 3.5) |
0.0 (Δ = + 3.5) |
0.1 (Δ = + 3.6) |
−3.4 (Δ = + 0.1) |
||||
| Morin | 35.7 | 44.7 (Δ = + 9.0) |
32.6 (Δ = −3.1) |
0.0 (Δ = −35.7) |
34.4 (Δ = −1.3) |
33.2 (Δ = −2.5) |
|||
| Myricetin | −9.1 | −10.1 (Δ = −1.0) |
−2.5 (Δ = + 6.6) |
−5.3 (Δ = + 3.8) |
0.0 (Δ = + 9.1) |
−6.3 (Δ = + 2.8) |
−7.7 (Δ = + 1.4) |
||
| Quercetin | −8.5 | −10.5 (Δ = −2.0) |
0.0 (Δ = + 8.5) |
0.0 (Δ = + 8.5) |
−5.9 (Δ = + 2.6) |
−5.7 (Δ = + 2.8) |
|||
| Flavonoid | Bond Dissociation Enthalpy | |||||||
|---|---|---|---|---|---|---|---|---|
| C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | ||
| Flavones | Acacetin | 102.1 | 91.1 | |||||
| Apigenin | 89.3 | 100.5 | 91.7 | |||||
| Chrysin | 102.1 | 92.8 | ||||||
| Chrysoeriol | 87.0 | 106.7 | 91.6 | |||||
| Diosmetin | 87.8 | 106.8 | 92.0 | |||||
| Genkwanin | 88.8 | 102.2 | ||||||
| Luteolin | 88.2 | 84.4 | 106.1 | 91.8 | ||||
| Flavonols | Fisetin | 83.4 | 85.1 | 86.7 | 90.3 | |||
| Galangin | 86.9 | 98.5 | 91.7 | |||||
| Kaempferol | 86.7 | 85.5 | 98.3 | 90.3 | ||||
| Morin | 93.4 | 88.8 | 84.6 | 99.1 | 91.4 | |||
| Myricetin | 87.5 | 80.7 | 84.4 | 86.1 | 98.3 | 91.0 | ||
| Quercetin | 87.4 | 81.4 | 85.5 | 98.0 | 90.4 | |||
| Flavonoid | Ionization Potential | Proton Dissociation Enthalpy | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | |||
| Flavones | Acacetin | 115.9 | 16.1 | 5.0 | |||||
| Apigenin | 117.0 | 2.2 | 13.4 | 4.6 | |||||
| Chrysin | 120.5 | 11.4 | 2.1 | ||||||
| Chrysoeriol | 113.4 | 3.5 | 23.2 | 8.1 | |||||
| Diosmetin | 113.8 | 3.9 | 22.9 | 8.1 | |||||
| Genkwanin | 117.2 | 1.4 | 14.8 | ||||||
| Luteolin | 115.0 | 3.1 | −0.7 | 20.9 | 6.6 | ||||
| Flavonols | Fisetin | 108.9 | 4.4 | 6.0 | 7.6 | 11.3 | |||
| Galangin | 114.3 | 2.4 | 14.0 | 7.2 | |||||
| Kaempferol | 110.2 | 6.4 | 5.2 | 18.0 | 10.0 | ||||
| Morin | 113.3 | 10.0 | 5.4 | 1.3 | 15.7 | 8.1 | |||
| Myricetin | 109.0 | 8.4 | 1.5 | 5.3 | 6.9 | 19.2 | 11.9 | ||
| Quercetin | 108.9 | 8.4 | 2.4 | 6.5 | 19.0 | 11.4 | |||
| Flavonoid | Proton Affinity Enthalpy | Electron Transfer Enthalpy | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | ||
| Flavones | Acacetin | 45.0 | 30.8 | 87.0 | 90.2 | ||||||||||
| Apigenin | 31.8 | 43.3 | 31.4 | 87.4 | 87.1 | 90.1 | |||||||||
| Chrysin | 44.3 | 31.0 | 87.7 | 91.7 | |||||||||||
| Chrysoeriol | 33.3 | 45.0 | 30.7 | 83.5 | 91.6 | 90.8 | |||||||||
| Diosmetin | 36.7 | 45.3 | 30.7 | 81.0 | 91.4 | 91.1 | |||||||||
| Genkwanin | 31.1 | 46.0 | 87.5 | 86.0 | |||||||||||
| Luteolin | 35.8 | 27.4 | 43.8 | 31.3 | 82.3 | 86.8 | 92.1 | 90.3 | |||||||
| Flavonols | Fisetin | 30.7 | 34.1 | 38.5 | 30.0 | 82.6 | 80.8 | 78.1 | 90.2 | ||||||
| Galangin | 35.2 | 40.8 | 30.3 | 81.6 | 87.6 | 91.3 | |||||||||
| Kaempferol | 32.0 | 35.5 | 42.0 | 30.1 | 84.6 | 79.8 | 86.2 | 90.1 | |||||||
| Morin | 39.2 | 32.1 | 28.3 | 40.6 | 29.5 | 84.0 | 86.6 | 86.2 | 88.3 | 91.8 | |||||
| Myricetin | 36.0 | 29.1 | 30.1 | 34.9 | 41.2 | 29.9 | 81.3 | 81.5 | 84.2 | 81.0 | 87.0 | 90.9 | |||
| Quercetin | 36.7 | 27.2 | 35.5 | 41.5 | 30.0 | 80.6 | 84.0 | 79.8 | 86.4 | 90.3 | |||||
| Flavonoid | Reorganization Enthalpies | Hydrogen Abstraction Enthalpies | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | C2’ | C3’ | C4’ | C5’ | C3 | C5 | C7 | ||
| Flavones | Acacetin | −8.7 | −5.1 | 93.5 | 86.0 | ||||||||||
| Apigenin | −6.7 | −7.1 | −5.0 | 82.6 | 93.5 | 86.7 | |||||||||
| Chrysin | −8.6 | −5.6 | 93.5 | 87.2 | |||||||||||
| Chrysoeriol | −7.7 | −2.4 | −5.2 | 79.3 | 104.3 | 86.5 | |||||||||
| Diosmetin | −7.2 | −2.4 | −5.4 | 80.6 | 104.4 | 86.6 | |||||||||
| Genkwanin | −6.3 | −8.5 | 82.4 | 93.7 | |||||||||||
| Luteolin | −7.0 | −8.6 | −1.7 | −5.0 | 81.2 | 75.9 | 104.4 | 86.8 | |||||||
| Flavonols | Fisetin | −7.5 | −7.8 | −9.1 | −5.9 | 75.9 | 77.3 | 77.6 | 84.4 | ||||||
| Galangin | −8.6 | −8.4 | −5.8 | 78.3 | 90.1 | 85.9 | |||||||||
| Kaempferol | −6.9 | −8.7 | −8.5 | −5.5 | 79.8 | 76.8 | 89.8 | 84.8 | |||||||
| Morin | −8.4 | −6.2 | −12.6 | −9.1 | −5.3 | 85.0 | 82.6 | 72.1 | 90.0 | 86.1 | |||||
| Myricetin | −6.8 | −8.3 | −7.2 | −9.4 | −8.5 | −6.0 | 80.7 | 72.4 | 77.1 | 76.7 | 89.8 | 85.0 | |||
| Quercetin | −7.0 | −8.0 | −8.8 | −8.7 | −5.6 | 80.4 | 73.4 | 76.7 | 89.3 | 84.9 | |||||
| Activity | HAT | ETPT | SPLET |
|---|---|---|---|
 |
Myricetin | Fisetin, Quercetin | Quercetin |
| Quercetin | Luteolin | ||
| Fisetin | Myricetin | Morin | |
| Luteolin | Kaempferol | Mirycetin | |
| Morin | Morin | Fisetin | |
| Kaempferol | Chrysoeriol | Kaempferol | |
| Galangin | Diosmetin | Galangin | |
| Chrysoeriol | Galangin | Chrysoeriol, Diosmetin | |
| Diosmetin | Luteolin | ||
| Genkanin | Acacetin | Acacetin | |
| Apigenin | Apigenin | Chrysine | |
| Accacetin | Genkwanin | Genkwanin | |
| Chrysine | Chrysine | Apigenin |
| Flavonoid | HAT | ETPT | SPLET | |||
|---|---|---|---|---|---|---|
| BDE | IP | PDE | PA | ETE | ||
| Luteolin | 103.6 | −282.6 | 416.1 | 26.4 | 107.0 | |
| Fisetin | 91.6 | −287.8 | 409.2 | 17.1 | 104.3 | |
| Myricetin | C3’–C4’ | 90.5 | −284.9 | 405.3 | 19.7 | 100.6 |
| C4’–C5’ | 83.7 | 398.5 | 15.5 | 98.1 | ||
| Quercetin | 93.8 | −285.7 | 409.4 | 18.5 | 105.2 | |
| Flavonoid | ΔrG0 | o-HFE | RE | Σ | |
|---|---|---|---|---|---|
| Luteolin | −5.0 | −5.6 | −34.4 | −40.0 | |
| Fisetin | −13.2 | −14.4 | −15.7 | 30.1 | |
| Myricetin | C3’–C4’ | −11.9 | −11.9 | −14.4 | −26.3 |
| Myricetin | C4’–C5’ | −7.5 | −8.2 | −13.8 | −22.0 |
| Quercetin | 15.1 | −14.4 | −15.7 | −30.1 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spiegel, M.; Andruniów, T.; Sroka, Z. Flavones’ and Flavonols’ Antiradical Structure–Activity Relationship—A Quantum Chemical Study. Antioxidants 2020, 9, 461. https://doi.org/10.3390/antiox9060461
Spiegel M, Andruniów T, Sroka Z. Flavones’ and Flavonols’ Antiradical Structure–Activity Relationship—A Quantum Chemical Study. Antioxidants. 2020; 9(6):461. https://doi.org/10.3390/antiox9060461
Chicago/Turabian StyleSpiegel, Maciej, Tadeusz Andruniów, and Zbigniew Sroka. 2020. "Flavones’ and Flavonols’ Antiradical Structure–Activity Relationship—A Quantum Chemical Study" Antioxidants 9, no. 6: 461. https://doi.org/10.3390/antiox9060461