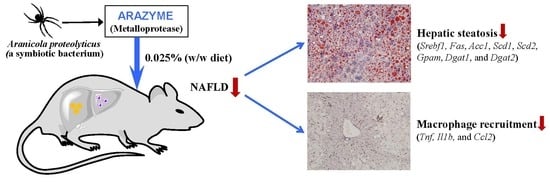
Arazyme Suppresses Hepatic Steatosis and Steatohepatitis in Diet-Induced Non-Alcoholic Fatty Liver Disease-Like Mouse Model
Abstract
:1. Introduction
2. Results
2.1. Effects of Arazyme on Metabolic Parameters
2.2. Effects of Arazyme on Plasma Biomarkers
2.3. Arazyme Reduced HFD-Induced Hepatic Steatosis
2.4. Arazyme Inhibited Lipid Accumulation in HepG2 Cells
2.5. Arazyme Suppressed Steatohepatitis and Macrophage Recruitment
2.6. Arazyme Inhibited PA-Induced Inflammation in RAW264.7 Macrophages
3. Discussion
4. Materials and Methods
4.1. Animals and Diets
4.2. Measurement of Metabolic Parameters
4.3. Histological Analysis of the Liver
4.4. Real-Time Quantitative RT-PCR (qRT-PCR)
4.5. Western Blotting Analysis
4.6. Experiments in HepG2 Cells and RAW264.7 Cells
4.7. Statistical Analyses
Author Contributions
Funding
Conflicts of Interest
References
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human fatty liver disease: Old questions and new insights. Science 2011, 332, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Joshi-Barve, S.; Barve, S.S.; Amancherla, K.; Gobejishvili, L.; Hill, D.; Cave, M.; Hote, P.; McClain, C.J. Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology 2007, 46, 823–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiniakos, D.G.; Vos, M.B.; Brunt, E.M. Nonalcoholic fatty liver disease: Pathology and pathogenesis. Annu. Rev. Pathol. 2010, 5, 145–171. [Google Scholar] [CrossRef] [PubMed]
- Marchesini, G.; Brizi, M.; Bianchi, G.; Tomassetti, S.; Bugianesi, E.; Lenzi, M.; McCullough, A.J.; Natale, S.; Forlani, G.; Melchionda, N. Nonalcoholic fatty liver disease: A feature of the metabolic syndrome. Diabetes 2001, 50, 1844–1850. [Google Scholar] [CrossRef]
- Alakhali, M.S.; Al-Maweri, S.A.; Al-Shamiri, H.M.; Al-Haddad, K.; Halboub, E. The potential association between periodontitis and non-alcoholic fatty liver disease: A systematic review. Clin. Oral Investig. 2018, 22, 2965–2974. [Google Scholar] [CrossRef] [PubMed]
- Churchward-Venne, T.A.; Pinckaers, P.J.M.; van Loon, J.J.A.; van Loon, L.J.C. Consideration of insects as a source of dietary protein for human consumption. Nutr. Rev. 2017, 75, 1035–1045. [Google Scholar] [CrossRef]
- Nowak, V.; Persijn, D.; Rittenschober, D.; Charrondiere, U.R. Review of food composition data for edible insects. Food Chem. 2016, 193, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Rumpold, B.A.; Schluter, O.K. Nutritional composition and safety aspects of edible insects. Mol. Nutr. Food Res. 2013, 57, 802–823. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.A.; Jeong, T.S.; Park, D.S.; Xu, M.Z.; Oh, H.W.; Song, K.B.; Lee, W.S.; Park, H.Y. Antioxidant effects of quinoline alkaloids and 2,4-di-tert-butylphenol isolated from Scolopendra subspinipes. Biol. Pharm. Bull. 2006, 29, 735–739. [Google Scholar] [CrossRef]
- Xu, M.Z.; Lee, W.S.; Han, J.M.; Oh, H.W.; Park, D.S.; Tian, G.R.; Jeong, T.S.; Park, H.Y. Antioxidant and anti-inflammatory activities of N-acetyldopamine dimers from Periostracum Cicadae. Bioorg. Med. Chem. 2006, 14, 7826–7834. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, Y.S.; Moon, H.I.; Cho, Y.S. Hepatoprotective effects on alcoholic liver disease of fermented silkworms with Bacillus subtilis and Aspergillus kawachii. Int. J. Food Sci. Nutr. 2012, 63, 537–547. [Google Scholar] [CrossRef]
- Seo, M.; Goo, T.W.; Chung, M.Y.; Baek, M.; Hwang, J.S.; Kim, M.A.; Yun, E.Y. Tenebrio molitor larvae inhibit adipogenesis through AMPK and MAPKs signaling in 3T3-L1 adipocytes and obesity in high-fat diet-induced obese mice. Int. J. Mol. Sci. 2017, 18, e518. [Google Scholar] [CrossRef]
- Bersanetti, P.A.; Park, H.-Y.; Bae, K.S.; Son, K.-H.; Shin, D.-H.; Hirata, I.Y.; Juliano, M.A.; Carmona, A.K.; Juliano, L. Characterization of arazyme, an exocellular metalloprotease isolated from Serratia proteamaculans culture medium. Enzym. Microb. Technol. 2005, 37, 574–581. [Google Scholar] [CrossRef]
- Kwak, J.; Lee, K.; Shin, D.H.; Maeng, J.S.; Park, D.S.; Oh, H.W.; Son, K.H.; Bae, K.S.; Park, H.Y. Biochemical and genetic characterization of arazyme, an extracellular metalloprotease produced from Serratia proteamaculans HY-3. J. Microbiol. Biotechnol. 2007, 17, 761–768. [Google Scholar]
- Beuth, J. Proteolytic enzyme therapy in evidence-based complementary oncology: Fact or fiction? Integr. Cancer Ther. 2008, 7, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Yang, E.J.; Shin, D.H.; Son, K.H.; Park, H.Y.; Lee, J.S. Effect of arazyme on the lipopolysaccharideinduced inflammatory response in human endothelial cells. Mol. Med. Rep. 2014, 10, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Kim, M.J.; Shin, D.H.; Son, K.H.; Park, H.Y.; Lee, J.S. Arazyme inhibits cytokine expression and upregulates skin barrier protein expression. Mol. Med. Rep. 2013, 8, 551–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, I.S.; Lee, N.R.; Baek, S.Y.; Kim, E.J.; Kim, J.S.; Jeong, T.S.; Shin, D.H.; Park, H.Y.; Lee, J.S. Inhibitory effect of arazyme on the development of atopic dermatitis-like lesions in BALB/c and Nc/Nga mice. Mol. Med. Rep. 2015, 11, 3995–4001. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Jeong, D.H.; Park, H.Y.; Son, K.H.; Shin, D.H.; Do, S.H.; Yang, H.J.; Yuan, D.W.; Hong, I.H.; Goo, M.J.; et al. Hepatoprotective effect of Arazyme on CCl4-induced acute hepatic injury in SMP30 knock-out mice. Toxicology 2008, 246, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Imajo, K.; Yoneda, M.; Kessoku, T.; Ogawa, Y.; Maeda, S.; Sumida, Y.; Hyogo, H.; Eguchi, Y.; Wada, K.; Nakajima, A. Rodent models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Int. J. Mol. Sci. 2013, 14, 21833–21857. [Google Scholar] [CrossRef]
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 2002, 109, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Shah, N.A.; Warrington, J.A.; Anderson, N.N.; Park, S.W.; Brown, M.S.; Goldstein, J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 2003, 100, 12027–12032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iizuka, K.; Bruick, R.K.; Liang, G.; Horton, J.D.; Uyeda, K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA 2004, 101, 7281–7286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, H.; Takenoshita, M.; Sakurai, M.; Bruick, R.K.; Henzel, W.J.; Shillinglaw, W.; Arnot, D.; Uyeda, K. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA 2001, 98, 9116–9121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef]
- Xu, X.; So, J.S.; Park, J.G.; Lee, A.H. Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP. Semin. Liver Dis. 2013, 33, 301–311. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Feng, D.; Wang, Q.; Abdulla, A.; Xie, X.J.; Zhou, J.; Sun, Y.; Yang, E.S.; Liu, L.P.; Vaitheesvaran, B.; et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Investig. 2012, 122, 2417–2427. [Google Scholar] [CrossRef]
- Jelenik, T.; Kaul, K.; Sequaris, G.; Flogel, U.; Phielix, E.; Kotzka, J.; Knebel, B.; Fahlbusch, P.; Horbelt, T.; Lehr, S.; et al. Mechanisms of insulin resistance in primary and secondary nonalcoholic fatty liver. Diabetes 2017, 66, 2241–2253. [Google Scholar] [CrossRef]
- Brunt, E.M.; Tiniakos, D.G. Histopathology of nonalcoholic fatty liver disease. World J. Gastroenterol. 2010, 16, 5286–5296. [Google Scholar] [CrossRef]
- Tacke, F. Targeting hepatic macrophages to treat liver diseases. J. Hepatol. 2017, 66, 1300–1312. [Google Scholar] [CrossRef]
- Heymann, F.; Tacke, F. Immunology in the liver--from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 88–110. [Google Scholar] [CrossRef]
- Miura, K.; Yang, L.; van Rooijen, N.; Ohnishi, H.; Seki, E. Hepatic recruitment of macrophages promotes nonalcoholic steatohepatitis through CCR2. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1310–G1321. [Google Scholar] [CrossRef] [Green Version]
- Bekaert, M.; Verhelst, X.; Geerts, A.; Lapauw, B.; Calders, P. Association of recently described adipokines with liver histology in biopsy-proven non-alcoholic fatty liver disease: A systematic review. Obes. Rev. 2016, 17, 68–80. [Google Scholar] [CrossRef]
- Tacke, F.; Zimmermann, H.W. Macrophage heterogeneity in liver injury and fibrosis. J. Hepatol. 2014, 60, 1090–1096. [Google Scholar] [CrossRef] [Green Version]
- Pradere, J.P.; Kluwe, J.; De Minicis, S.; Jiao, J.J.; Gwak, G.Y.; Dapito, D.H.; Jang, M.K.; Guenther, N.D.; Mederacke, I.; Friedman, R.; et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 2013, 58, 1461–1473. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, Z.; Klaunig, J.E. Modulation of xenobiotic nuclear receptors in high-fat diet induced non-alcoholic fatty liver disease. Toxicology 2018, 410, 199–213. [Google Scholar] [CrossRef]
- Pereira, F.V.; Ferreira-Guimaraes, C.A.; Paschoalin, T.; Scutti, J.A.; Melo, F.M.; Silva, L.S.; Melo, A.C.; Silva, P.; Tiago, M.; Matsuo, A.L.; et al. A natural bacterial-derived product, the metalloprotease arazyme, inhibits metastatic murine melanoma by inducing MMP-8 cross-reactive antibodies. PLoS ONE 2014, 9, e96141. [Google Scholar] [CrossRef]
- Van Lint, P.; Wielockx, B.; Puimege, L.; Noel, A.; Lopez-Otin, C.; Libert, C. Resistance of collagenase-2 (matrix metalloproteinase-8)-deficient mice to TNF-induced lethal hepatitis. J. Immunol. 2005, 175, 7642–7649. [Google Scholar] [CrossRef]
- Baig, M.S.; Yaqoob, U.; Cao, S.; Saqib, U.; Shah, V.H. Non-canonical role of matrix metalloprotease (MMP) in activation and migration of hepatic stellate cells (HSCs). Life Sci. 2016, 155, 155–160. [Google Scholar] [CrossRef]
- Shaker, E.; Mahmoud, H.; Mnaa, S. Silymarin, the antioxidant component and Silybum marianum extracts prevent liver damage. Food Chem. Toxicol. 2010, 48, 803–806. [Google Scholar] [CrossRef]
- Ni, X.; Wang, H. Silymarin attenuated hepatic steatosis through regulation of lipid metabolism and oxidative stress in a mouse model of nonalcoholic fatty liver disease (NAFLD). Am. J. Transl. Res. 2016, 8, 1073–1081. [Google Scholar]
- Li, H.; Kim, U.H.; Yoon, J.H.; Ji, H.S.; Park, H.M.; Park, H.Y.; Jeong, T.S. Suppression of hyperglycemia and hepatic steatosis by black-soybean-leaf extract via enhanced adiponectin-receptor signaling and AMPK activation. J. Agric. Food Chem. 2019, 67, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yoon, J.H.; Won, H.J.; Ji, H.S.; Yuk, H.J.; Park, K.H.; Park, H.Y.; Jeong, T.S. Isotrifoliol inhibits pro-inflammatory mediators by suppression of TLR/NF-κB and TLR/MAPK signaling in LPS-induced RAW264.7 cells. Int. Immunopharmacol. 2017, 45, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Jeong, J.M.; Kim, S.J.; Seo, W.; Kim, M.H.; Choi, W.M.; Yoo, W.; Lee, J.H.; Shim, Y.R.; Yi, H.S.; et al. Pro-inflammatory hepatic macrophages generate ROS through NADPH oxidase 2 via endocytosis of monomeric TLR4-MD2 complex. Nat. Commun. 2017, 8, 2247. [Google Scholar] [CrossRef] [PubMed]








| ND | HFD | HFD+Ara | HFD+MT | |
|---|---|---|---|---|
| Initial body weight (g) | 23.5 ± 0.3 | 23.6 ± 0.3 | 23.3 ± 0.4 | 23.5 ± 0.4 |
| Final body weight (g) | 38.8 ± 0.8 ‡ | 44.9 ± 1.4 ‡, ## | 44.4 ± 0.8 ‡ | 43.6 ± 1.9 ‡ |
| Weight gain (g) | 15.5 ± 0.5 | 21.3 ± 1.2 ## | 21.5 ± 0.7 | 20.2 ± 1.5 |
| Food intake (g/day) | 2.3 ± 0.1 | 2.0 ± 0.2 # | 2.0 ± 0.1 | 2.1 ± 0.1 |
| Liver (g) | 1.28 ± 0.04 | 1.36 ± 0.09 | 1.29 ± 0.06 | 1.35 ± 0.13 |
| ND | HFD | HFD+Ara | HFD+MT | |
|---|---|---|---|---|
| Glucose | ||||
| Initial (mg/dL) | 99.9 ± 3.5 | 100.6 ± 3.7 | 99.5 ± 2.6 | 100.9 ± 4.3 |
| Final (mg/dL) | 126.4 ± 10.3 † | 169.0 ± 12.9 ‡, # | 145.8 ± 7.9 ‡ | 152.5 ± 14.7 ‡ |
| Insulin | ||||
| Initial (ng/mL) | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.5 ± 0.1 |
| Final (ng/mL) | 0.9 ± 0.2 ‡ | 2.3 ± 0.5 ‡, # | 2.5 ± 0.4 ‡ | 2.3 ± 0.4 ‡ |
| HbA1c (%) | 5.05 ± 0.09 | 5.41 ± 0.12 # | 5.15 ± 0.06 | 5.12 ± 0.13 * |
| HOMA-IR | 6.9 ± 1.3 | 19.1 ± 4.7 # | 16.4 ± 3.0 | 17.5 ± 4.3 |
| TG (mg/dL) | 118.2 ± 7.5 | 142.7 ± 9.5 # | 118.4 ± 4.2 * | 111.7 ± 10.0 ** |
| TC (mg/dL) | 144.5 ± 6.4 | 183.1 ± 9.4 # | 170.6 ± 7.2 | 162.0 ± 13.0 |
| HDL-C (mg/dL) | 74.4 ± 6.0 | 73.2 ± 2.6 | 68.0 ± 2.2 | 63.7 ± 3.6 |
| HDL-C/TC (%) | 50.7 ± 3.5 | 40.9 ± 2.0 ## | 39.7 ± 1.5 | 40.4 ± 1.6 |
| NEFA (mEq/L) | 2.1 ± 0.1 | 2.7 ± 0.1 # | 2.3 ± 0.1 * | 2.2 ± 0.2 * |
| AST (IU/L) | 96.5 ± 6.1 | 117.4 ± 5.8 # | 105.8 ± 11.3 | 111.8 ± 10.4 |
| ALT (IU/L) | 26.0 ± 3.6 | 45.3 ± 10.2 # | 41.5 ± 5.7 | 40.2 ± 4.2 |
| Gene (Number) | Forward Primer | Reverse Primer |
|---|---|---|
| Acc1 (NM_133360) | AGTTTCCCAGCCAGCAGATT | ATCCATCACCACAGCCTTCA |
| Acc2 (NM_133904) | CCCATCACCACTCCTTCTGA | GTCCGAGTCTCCACAGCAAT |
| Acox (NM_015729) | GCTGGGCTGAAGGCTTTTACTA | AATCCCACTGCTGTGAGAATAGC |
| Ccl2 (NM_011333) | TCACCTGCTGCTACTCATTC | TACAGAAGTGCTTGAGGTGG |
| Ccl3 (NM_011337) | CACCCTCTGTCACCTGCTCAA | TGGCGCTGAGAAGACTTGGT |
| Ccl4 (NM_013652) | CTAACCCCGAGCAACACCAT | AGCCCATTGGTGCTGAGAAC |
| Ccl5 (NM_013653) | TCCCTGTCATTGCTTGCTCTAG | GAGCAGCTGAGATGCCCATT |
| Cpt1α (NM_013495) | CTGCACTCCTGGAAGAAGAA | GTTCTTCGTCTGGCTTGACA |
| Dgat1 (NM_010046) | ACAACCTGACCTACCGAGAT | AGTAGGGACCATCCACTGTT |
| Dgat2 (NM_026384) | GCTGGCATTTGACTGGAACA | TGGTCAGCAGGTTGTGTGTCTT |
| Fas (NM_007988) | TGTGAGTGGTTCAGAGGCAT | TTCTGTAGTGCCAGCAAGCT |
| Gapdh (NM_001001303) | ACATCATCCCTGCATCCACT | AGATCCACGACGGACACATT |
| Gpam (NM_001356285) | CCGGAAGAGGCCCTTCGTGG | GCCAGCCATCCTCTGTGCCT |
| Il1b (NM_008361) | ATGGCAACTGTTCCTGAACTCAACT | ATATTCTGTCCATTGAGGTGGAGAGCT |
| Mlxipl (NM_021455) | CAGATGCGGGACATGTTTGA | AATAAAGGTCGGATGAGGATGCT |
| Pparα (NM_011144) | CCTGAACATCGAGTGTCGAA | GTACTGGCATTTGTTCCGGT |
| Scd1 (NM_009127) | ACGCCGACCCTCACAATTC | AGTTTTCCGCCCTTCTCTTTG |
| Scd2 (NM_009128) | CCGTGGCTTCTTTTTCTCTCA | TTCCGCCCTTCTCTTTGACA |
| Srebf1 (NM_011480) | GAGCGAGCGTTGAACTGTAT | ATGCTGGAGCTGACAGAGAA |
| Srebf2 (NM_033218) | TCCTCCATCAACGACAAAATCA | ACTTGTGCATCTTGGCATCTGT |
| Tnfa (NM_001278601) | CTCAGATCATCTTCTCAAAATTCGAGTGACA | CTTCACAGAGCAATGACTCCAAAGT |
| Ucp2 (NM_011671) | GCCTCTACGACTCTGTCAAA | CTTCGACAGTGCTCTGGTAT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Yoo, W.; Park, H.-M.; Lim, S.-Y.; Shin, D.-H.; Kim, S.; Park, H.-Y.; Jeong, T.-S. Arazyme Suppresses Hepatic Steatosis and Steatohepatitis in Diet-Induced Non-Alcoholic Fatty Liver Disease-Like Mouse Model. Int. J. Mol. Sci. 2019, 20, 2325. https://doi.org/10.3390/ijms20092325
Li H, Yoo W, Park H-M, Lim S-Y, Shin D-H, Kim S, Park H-Y, Jeong T-S. Arazyme Suppresses Hepatic Steatosis and Steatohepatitis in Diet-Induced Non-Alcoholic Fatty Liver Disease-Like Mouse Model. International Journal of Molecular Sciences. 2019; 20(9):2325. https://doi.org/10.3390/ijms20092325
Chicago/Turabian StyleLi, Hua, Wonbeak Yoo, Hye-Mi Park, Soo-Youn Lim, Dong-Ha Shin, Seokho Kim, Ho-Yong Park, and Tae-Sook Jeong. 2019. "Arazyme Suppresses Hepatic Steatosis and Steatohepatitis in Diet-Induced Non-Alcoholic Fatty Liver Disease-Like Mouse Model" International Journal of Molecular Sciences 20, no. 9: 2325. https://doi.org/10.3390/ijms20092325