-
PDF
- Split View
-
Views
-
Cite
Cite
Paul D. Pennington, Liliana M. Costa, Jose F. Gutierrez-Marcos, Andy J. Greenland, Hugh G. Dickinson, When Genomes Collide: Aberrant Seed Development Following Maize Interploidy Crosses, Annals of Botany, Volume 101, Issue 6, April 2008, Pages 833–843, https://doi.org/10.1093/aob/mcn017
Close - Share Icon Share
Abstract
The results of wide- or interploidy crosses in angiosperms are unpredictable and often lead to seed abortion. The consequences of reciprocal interploidy crosses have been explored in maize in detail, focusing on alterations to tissue domains in the maize endosperm, and changes in endosperm-specific gene expression.
Following reciprocal interploidy crosses between diploid and tetraploid maize lines, development of endosperm domains was studied using GUS reporter lines, and gene expression in resulting kernels was investigated using semi-quantitative RT-PCR on endosperms isolated at different stages of development.
Reciprocal interploidy crosses result in very small, largely infertile seeds with defective endosperms. Seeds with maternal genomic excess are smaller than those with paternal genomic excess, their endosperms cellularize earlier and they accumulate significant quantities of starch. Endosperms from the reciprocal cross undergo an extended period of cell proliferation, and accumulate little starch. Analysis of reporter lines and gene expression studies confirm that functional domains of the endosperm are severely disrupted, and are modified differently according to the direction of the interploidy cross.
Interploidy crosses affect factors which regulate the balance between cell proliferation and cell differentiation within the endosperm. In particular, unbalanced crosses in maize affect transfer cell differentiation, and lead to the temporal deregulation of the ontogenic programme of endosperm development.
INTRODUCTION
Not only do two distinct haploid genomes fuse during fertilization in eukaryotes, but also two different populations of molecules regulating chromatin structure and function come together in the zygotic nucleus. In addition to regulatory proteins, both male and female nuclei bring with them their own sets of non-coding RNAs (Engel et al., 2003) with the capacity to control transcript levels and remodel chromatin. For this reason, any eukaryotic fertilization, apart perhaps from the conjunction of two haploid, genetically similar genomes, can produce unpredictable results. An ability to foretell the outcomes of wide crosses, and crosses between plants of different ploidy levels would be of great value to plant breeders and geneticists. However, tools to investigate molecular interactions comprising fertilization and its consequences in plants are in their infancy. Gene expression immediately following nuclear fusion has been followed in maize zygotes and endosperms, using isolated cells (Le et al., 2005), while Hegarty et al. (2006) employed ‘orphan arrays’ (containing largely unsequenced genes) to provide the first molecular survey of hybridization in Senecio. Although this represents a promising start to the molecular dissection of hybridization, other approaches are needed to unravel the gene networks that regulate early seed development following fertilization.
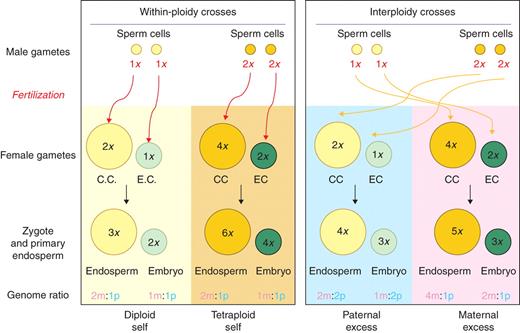
The outcomes of ‘normal’ diploid-by-diploid within-species pollinations are relatively predictable, but molecular analysis of these events is hampered by the lack of polymorphisms by which the activity of paternal and maternal genes can be determined. Furthermore, the very predictability of within-species fertilization is almost certainly the net result of evolution acting to ‘harmonize’ the chromatin-regulating systems described above. Adopting a practical approach to unravelling these interactions, we have focused on the fusion between the single sperm and two maternal nuclei that occurs in the central cell following double fertilization in maize and generates the endosperm (Lopes and Larkins, 1993; Becraft, 2001; Baroux et al., 2002; Berger, 2003; Costa et al., 2004; Olsen, 2004). This event already has an inbuilt genomic imbalance (two maternal to one paternal genomes) and, using reciprocal pollinations between inbred lines of different ploidy (2n × 4n and vice versa), we have examined the cell biological and molecular consequences of creating endosperms with maternal and paternal ‘genomic excess’ (Fig. 1). The outcomes of these interploidy crosses depend on a range of factors, including gene dosage (Birchler et al., 2005), genomic imprinting (Alleman and Doctor, 2000; Gehring et al., 2004) and the compatibility of the two sets of chromatin remodelling machinery discussed above (Dilkes and Comai, 2004; Madlung and Comai, 2004).
Schematic diagram showing consequences of same ploidy (‘balanced’) and interploidy (unbalanced) crosses in maize, showing the number – and parental origin (m, maternal; p, paternal) – of the haploid genomes present in the embryo and endosperm of each class of pollination.
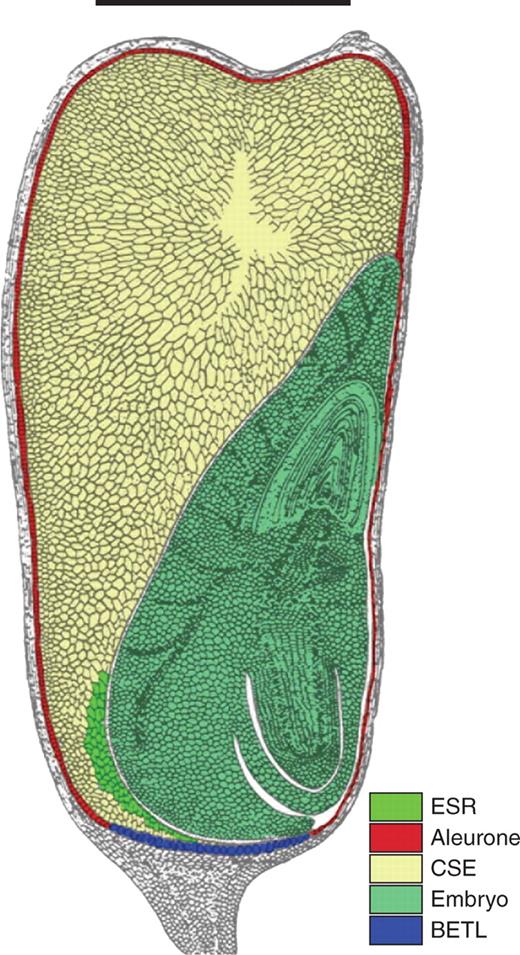
Maize endosperm comprises a number of functional domains or ‘tissues’ (Fig. 2). The central region, where starch is accumulated, is termed the central starchy endosperm (CSE) and is surrounded by a monolayer of epidermal cells, the aleurone, which plays an important part in seed germination (Lopes and Larkins, 1993; Walbot, 1994; Becraft, 2001). Nutrients from the maternal tissue enter through a group of specialized transfer cells [the basal endosperm transfer layer (BETL) (Thompson et al., 2001)], while evidence is accumulating of a specialized domain adjacent to the embryo [embryo surrounding region (ESR)], which may be involved in the exchange of resources and signals (Opsahl-Ferstad et al., 1997) The cytology of reciprocal interploidy crosses in maize was studied in detail by Cooper (1951) and Lin (1984) who reported that these pollinations result in aborted kernels, and that reciprocal crosses result in striking differences in embryo and endosperm development. This reciprocal asymmetry of endosperm development has been interpreted in terms of genomic imprinting (Scott et al., 1998), given that in some species the endosperm is known to be highly sensitive to departure from the two maternal, one paternal genomic balance (Lin, 1984). Cooper (1951) reported that the ‘adsorbing structure’ (presumably the BETL) was reduced in crosses with maternal genomic excess (MGE; three maternal genomes:one paternal genome), and Charlton et al. (1995) described modification of the BETL following pollinations involving paternal genomic excess (PGE; two maternal:two paternal genomes). More recently, a number of imprinted sequences have been shown to be expressed monoallelicly in the BETL, including MEG1, a putative component of a signalling pathway between the developing endosperm and the investing maternal tissue (Gutierrez-Marcos et al., 2004).
Diagram of mature maize kernel in longitudinal section. The embryo (light green) lies alongside the central starchy endosperm (CSE, light brown), which is surrounded by the aleurone (red), except in the region of the basal endosperm transfer layer (BETL, blue). The embryo surrounding region (ESR, green) is located at the base of the kernel, between the embryo and the CSE. Scale bar = 5 mm. (Adapted from Keisselbach, 1949.).
The present paper presents new data on maize grain development following reciprocal interploidy pollinations. Using GUS reporter constructs, early development of MGE and PGE endosperms is shown to be severely affected following interploidy pollinations, with key functional domains such as the aleurone and BETL being disrupted. Semi-quantitative RT-PCR analysis of genes defining endosperm domains revealed expression patterns that largely complement the histological data. The findings are consistent with a strong maternal control of early endosperm development and some of the predictions of the kinship (or ‘parental conflict’) hypothesis for plants (Haig and Westoby, 1991; Haig, 2000) and mammals (Hernandez et al., 2003), while others point to fundamental differences in gene regulation between MGE and PGE endosperms, particularly in later development.
MATERIALS AND METHODS
Plant material
Maize plants were grown to maturity in the greenhouse under a 16-h daylight cycle with supplementary lighting. Plants used in the study were as follows [seed origin shown square brackets]: To confirm the ploidy of individual lines, leaf material was analysed using flow cytometry (FACSScan; BD Biosciences, San Jose, CA, USA) as described by Dolezcaronel et al. (1998). Peaks were measured using CellQuest software (BD Biosciences). Pollinations were carried out as described in the Maize Handbook (Freeling and Walbot, 1996); the genomic consequences of balanced and interploidy crosses involving diploid and tetraploid lines are set out in Fig. 1.
Diploid inbred line: H99 [Maize Stock Centre].
Diploid marker lines: pBETL1::GUS (W23 BC4) [R. Thompson, INRA, Dijon, France]; pVp1::GUS (W23 BC4) [P. Perez, Biogemma SA, Clermont Ferrand, France].
Tetraploids: W23 [J. Gutierrez-Marcos, University of Oxford, UK].
Endosperm isolation, histology and microscopy
Seeds were stained for GUS activity and endosperms subsequently fixed, embedded, sectioned and stained according to Costa et al. (2003).
Semi-quantitative RT-PCR analysis
Endosperms were excised from kernels at different developmental stages by microdissection, and RNA was isolated and samples prepared as described in Le et al. (2005). Primers used for the amplification of endosperm sequences are listed in Pennington (2005). RT-PCR was carried out for 20, 25, 30 and 40 cycles.
RESULTS
Seed mass following balanced and unbalanced reciprocal interploidy crosses
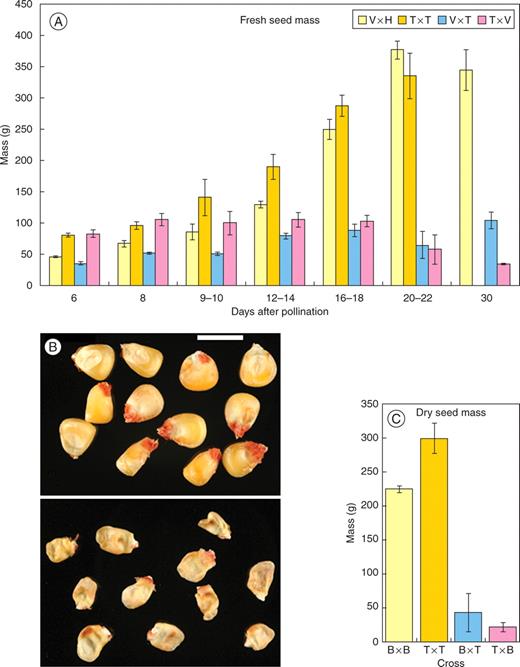
Fresh seed weights were recorded throughout development following balanced and interploidy crosses. Figure 3A shows change in seed mass over 25 d resulting from crosses between the pVp1::GUS (W23) line and the diploid inbred (H99) line, between the pVp1::GUS (W23) reporter and the tetraploid (W23) line, and from selfing the tetraploid (W23) plants. There is significant variation in these data, but the trends are clear; seeds from balanced diploid and tetraploid crosses accumulated mass rapidly between 10 and 20 d after pollination (DAP), while seeds from the interploidy crosses slowly gained in mass up to 12 DAP; thereafter, their fresh weight decreased to approx. 50 mg at 30 DAP compared with >300 mg of the balanced crosses.
Changes in seed mass during development following balanced and interploidy crosses. (A) Changes in seed weight of balanced and paternal and maternal genomic excess crosses involving the pVp1::GUS reporter line (v, pVp1::GUS line; H, H99 diploid inbred; T, tetraploid). Error bars show s.d. of the mean. (B) Mature dried kernels. Top, balanced cross (diploid selfed); bottom, paternal genomic excess cross (diploid × tetraploid). (C) Seed mass of mature desiccated kernels following balanced and interploidy crosses involving the diploid pBETL1::GUS reporter line (B) and the tetraploid (T). Error bars show s.d. of the mean.
The masses of MGE and PGE seed did not differ significantly during development (Fig. 3A), but the 30 DAP mean masses of seed formed from PGE crosses [H99(2n) × W23(4n)] were always greater than of seeds from the reciprocal pollination. This also occurred when similar studies were carried out with the pBETL1::GUS line (W23) as the diploid. A small proportion of the seeds were fertile, with viability seemingly dependent on the direction of the interploidy crosses. On average, 1·7% (N > 100) of seeds from PGE and 0·83% (N > 100) from MGE crosses germinated, pointing to a correlation between viability and seed weight [both fresh (Fig. 3A) and dry (Fig. 3B, C)]. The triploidy of the resulting seedlings was confirmed by using flow cytometry.
Activity of the pVP1::GUS reporter gene in balanced and unbalanced reciprocal interploidy crosses
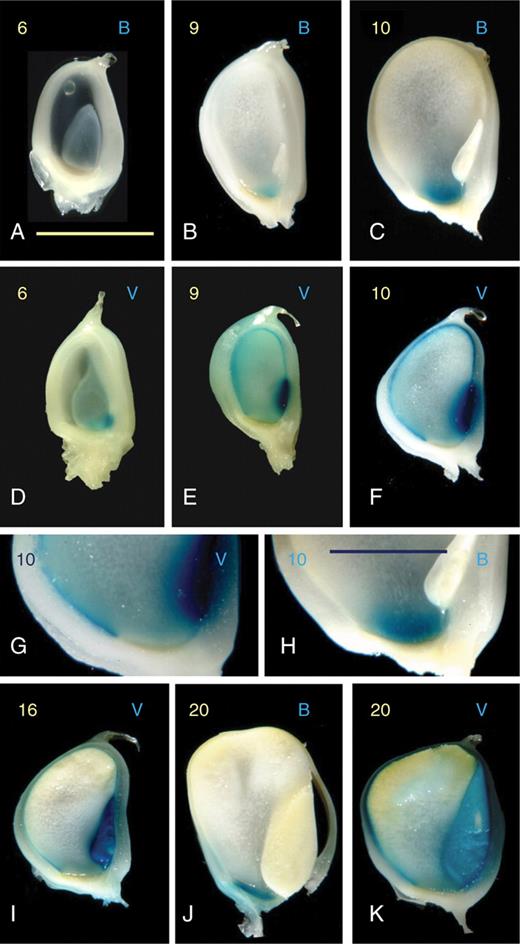
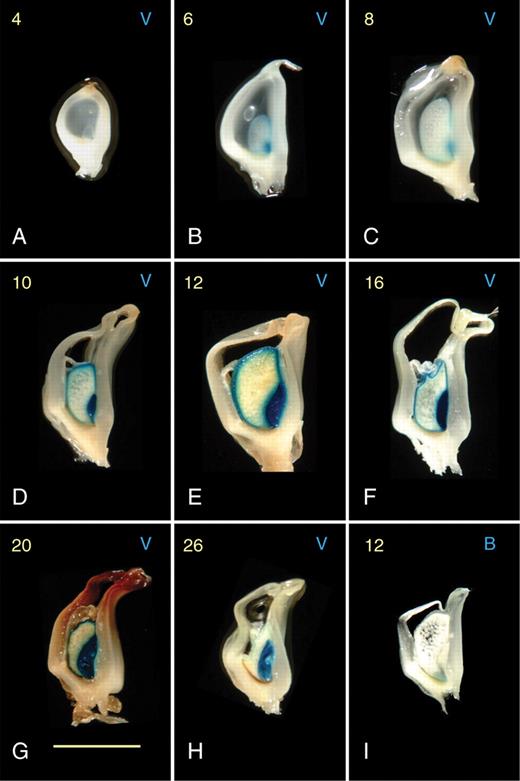
Aleurone development in seeds resulting from balanced crosses was investigated using the H99 diploid inbred and the pVp1::GUS and pBETL1::GUS transgenic reporter lines. Few differences were observed between reciprocal pollinations, and the distribution of signal observed was in agreement with a previous report (Costa et al., 2003). GUS activity was detectable in the embryo and developing aleurone layer from 6 DAP crosses (Fig. 4), and continued in these regions until approx. 24 DAP, when it became attenuated in the aleurone, but remained unchanged in the embryo. Interestingly, signal from the pVp1::GUS reporter at the endosperm periphery appeared at 5–7 DAP, some 3 d before the typical aleurone cell structure. pVp1::GUS signal was not seen in the BETL domain (Fig. 4).
Expression of the pBETL1::GUS and pVp1::GUS reporter lines in balanced crosses with H99 diploid inbred plants. Days after pollination (DAP) are shown in yellow on the top left of each frame, and the reporter line is indicated by B (pBETL::GUS) or V (pVp1::GUS) on the top right. Frames (A–F) and (I–K) show entire longitudinally sectioned kernels, (G) shows the absence of pVp1::GUS signal in the BETL region, and (H) signal in this domain from the pBETL1::GUS reporter. Scale bars: (A–F), (I–K), 5 µm; (G, H), 10 µm.
pVp1::GUS activity was studied between 4 and 26 DAP in MGE pollinations (Fig. 5). Signal first appeared only in the embryo at 4 DAP, 1 d sooner than in the control balanced cross. The distribution of GUS activity over the next 10 d resembled that of balanced crosses, except that kernel development was retarded. At 12 DAP the maternal-excess kernel closely resembled the control, except that signal was also formed in the BETL region (Fig. 5). Periodic acid-Schiffs (PAS) staining revealed that most of the cells in the ‘BETL domain’ had neither BETL nor aleurone morphology (data not shown). Between 15 and 30 DAP development became progressively more abnormal, with most of the endosperm degenerating, and the pVp1::GUS signal being restricted to the embryo, which by this stage occupied most of the kernel (Fig. 5).
Expression of the pBETL1::GUS and pVp1::GUS reporter lines in maternal genomic excess endosperms resulting from unbalanced crosses with tetraploid plants. Days after pollination (DAP; 4–26) are shown in yellow on the top left of each frame, and the reporter line is indicated by B (pBETL::GUS) or V (pVp1::GUS) on the top right. Only one example of a pBETL::GUS reporter line is shown, as signal was only visible 12 DAP. All frames show entire, longitudinally sectioned kernels. Scale bar = 5 µm.
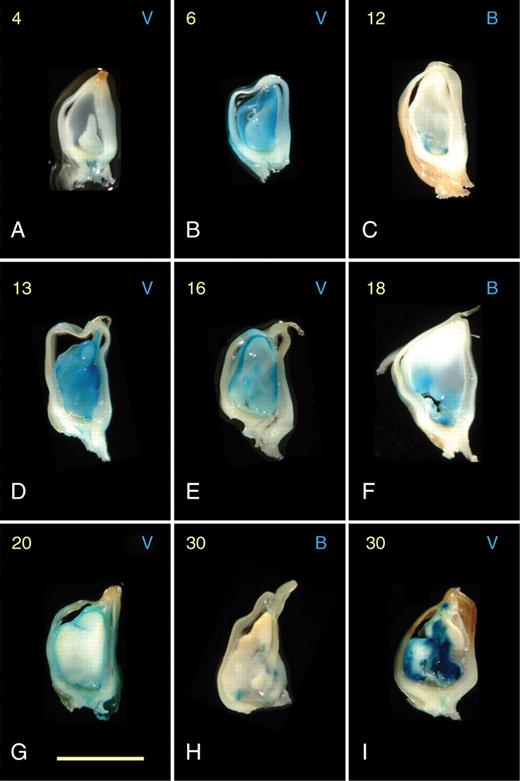
In PGE interploidy crosses between 4 and 26 DAP (Fig. 6), pVp1::GUS activity was first detected at 6–10 DAP, distributed generally throughout the endosperm but with higher levels at its germinal face and near the crown. However, following short incubation periods GUS activity was localized at the endosperm periphery, indicating diffusion of the GUS product during incubation. This migration of signal was not seen in balanced crosses, indicating structural differences in the interior of endosperms formed following balanced and PGE crosses. By 14 DAP, GUS activity had become more tightly focused at the periphery of the endosperm and, between 14 and 20 DAP the accumulation of starch resulted in a progressive loss of translucency in the central region. By this stage the endosperm surface had become convoluted and irregular, often with deep cavities penetrating the adgerminal face. The aleurone, following the contours of the endosperm surface, also appeared irregular and variable between kernels. The ‘ectopic’ surfaces generated by the fissures deep within the endosperm also exhibited GUS activity (Figs 6 and 7). By 30 DAP, high levels of signal were not only present at the endosperm periphery, but also in irregular ‘patches’ throughout the central region of the endosperm.
Expression of the diploid pBETL1::GUS and pVp1::GUS reporter lines in paternal excess endosperms resulting from unbalanced crosses with tetraploid plants. Days after pollination (DAP; 4–30) are shown in yellow on the top left of each frame, and the reporter line is indicated by B (pBETL::GUS) or V (pVp1::GUS) on the top right. All frames show entire, longitudinally sectioned kernels. Scale bar = 5 µm.
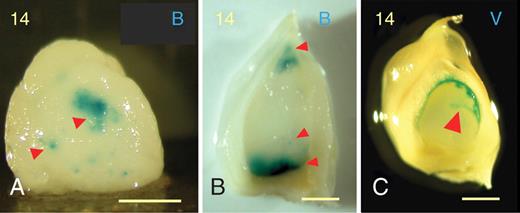
Kernels from paternal genomic excess crosses at 14 DAP showing ectopic expression (red arrowheads) of the pBETL1::GUS reporter (A, B), and expression (red arrowhead) of the pVp1::GUS construct in ‘fissures’ in the endosperm surface (C). Scale bars = 1 mm.
Activity of the pBETL1::GUS reporter gene in balanced and unbalanced reciprocal interploidy crosses
As previously reported (Hueros et al., 1999a), pBETL1::GUS transgene activity was detectable in the BETL region from 6 to 8 DAP in balanced crosses (Fig. 4). GUS expression in the reciprocal pollination was almost identical (data not shown). Comparison with PAS-stained material showed that GUS activity in the BETL domain at 5–6 DAP preceded formation of the specialized cell walls (Charlton et al., 1995; Davis and Smith, 1990; Thompson et al., 2001) by about 2 d.
Following MGE interploidy crosses, GUS activity was first detected later than in balanced cross controls (approx. 10–15 DAP) and was restricted to a smaller region, consistently located at the germinal face (Fig. 5). The strength of the GUS signal was weaker than in controls, and often undetectable. This expression pattern persisted throughout development, until approx. 15 DAP, when the signal attenuated immediately prior to the collapse of the endosperm. Sectioned kernels stained with PAS to reveal both cell-wall morphology and GUS activity showed a restricted group of cells with ‘normal’ BETL wall morphology in the region of the pBETL1::GUS reporter expression (data not shown).
Lines carrying the pBETL1::GUS transgene were pollinated with tetraploid plants to generate PGE endosperms, and GUS activity was investigated between 4 and 26 DAP (Fig. 6). Signal was first detected at 10 DAP, but instead of a continuous layer of signal between the endosperm and the nucellar tissue, small irregular groups of cells were seen to extend across this boundary. PAS staining of the BETL regions of all PGE endosperms showed that although cells differed morphologically from those in the central region and featured thicker cell walls, wall ingrowths characteristic of BETL cells were not present. As development progressed from 14 to 18 DAP, the groups of reporter-expressing cells increased in size and number until, in some endosperms, a near-continuous BETL was formed. Dramatic ectopic activity of the pBETL1::GUS reporter was also detected throughout the endosperm at this time-point (Figs 6 and 7), including expression at the apical tip (Fig. 7).
Altered levels of starch accumulation in endosperms following interploidy crosses
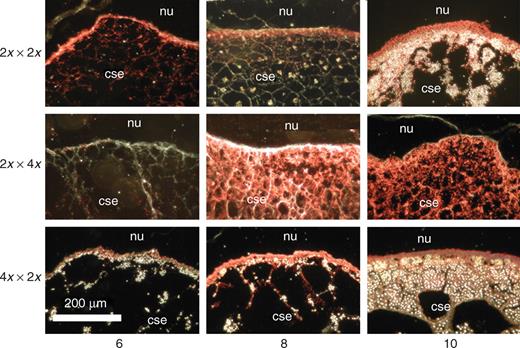
To compare levels of starch accumulation following balanced diploid pollinations and unbalanced interploidy crosses, dark-field microscopy was used to locate starch grains in developing endosperms. Starch was first detected at 6 DAP in MGE endosperms, compared with 8–10 DAP in diploid balanced controls (Fig. 8). In contrast, PGE endosperms showed delayed initiation of starch formation, grains being visible only from approx. 14 DAP.
Accumulation of starch following balanced and interploidy crosses (involving the pVp1::GUS reporter line) revealed by GUS signal and dark-field microscopy. Sections are from the germinal region of the kernel. Nature and direction of cross are shown in the left margin, and DAP at which the sample taken is given at the base of figure. GUS activity generates a red signal, and under dark-field conditions starch grains appear as white points. Nu, nucellus; cse, central starchy endosperm.
Early expression of genes defining endosperm domains following balanced and interploidy crosses
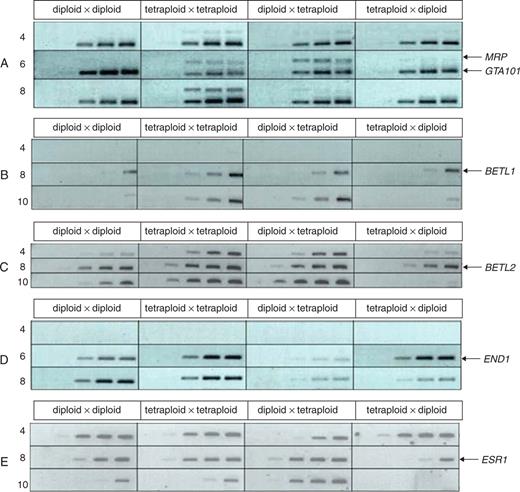
Semi-quantitative RT-PCR was used to investigate expression of genes defining endosperm domains isolated in endosperms from interploidy and balanced crosses at 4, 8 and 10 DAP. The genes analysed were: BETL-specific – BETL1,2, (Cai et al., 2002; Hueros et al., 1999a, b) and ZmMRP (Gomez et al., 2002); embryo surrounding region-specific – ZmESR1 (Opsahl-Ferstad et al., 1997); and basal endosperm – ZmEND1 (Gutierrez-Marcos et al., 2006b). GTA101 (maize transcription elongation factor, homologous to yeast SPT5) was used as a control.
Figure 9 shows that expression of the transfer layer marker BETL2 was down-regulated in MGE crosses at all time-points. BETL1 and BETL2 transcript levels were higher in the balanced tetraploid than in the balanced diploid control and, whereas the balanced tetraploid cross showed detectable levels of ZmMRP expression at all stages, the diploid control did not. Gene dosage may explain higher levels of expression in tetraploids, but the absence of ZmMRP transcript from diploids is perplexing and contrary to the reported expression pattern (Gomez et al., 2002); however, levels of this transcript were at the limits of detection in all endosperms. BETL1 is expressed more highly in PGE endosperms than in the diploid control, which reflects the generalized ectopic expression of the pBETL1::GUS transgene, and BETL2 is equally expressed following PGE and diploid crosses, with transcripts appearing earlier than those of BETL1. BETL2 is down-regulated in MGE endosperms. ZmEND1 levels are similar following diploid and MGE crosses, and PGE endosperms contain lower transcript levels. The expression pattern for ZmESR1 is similar in diploid and tetraploid controls, but in PGE endosperms fewer transcripts are present at 4 DAP than in the diploid and, following MGE pollinations, expression ceases altogether by 8 DAP.
RT-PCR analysis of endosperm gene expression from 4 to 10 DAP following balanced and reciprocal interploidy crosses involving the H99 diploid inbred, and the tetraploid lines. RT-PCR was carried out using 5 ng cDNA; individual bands represent PCR product taken after 20, 25, 30, 35 and 40 cycles. The nature and the direction of crosses are shown in the top line, and DAP in the left margin. (A) GTA101 (control) and ZmMRP (MRP); (B) BETL1; (C) BETL2; (D) ZmEND (END); (E) ZmESR1 (ESR1).
DISCUSSION
Maternal or paternal genomic excess results in seed abortion in maize
The regular abortion of seed development following interploidy pollinations in maize (Cooper, 1951; Charlton et al., 1995) is held to result from endosperm failure caused by ‘genomic imbalance’ in the endosperm (Lin, 1984; Gutierrez-Marcos et al., 2003). By contrast, interploidy pollinations succeed in Arabidopsis thaliana (Arabidopsis), with crosses between tetraploids and diploids being fully fertile. However, development of the seed is radically different depending on whether the genomic excess comes from the male or female parent (Scott et al., 1998). Paternal genomic excess results in larger seeds and maternal excess in smaller seeds in Arabidopsis. These differences result from an extended proliferation phase of endosperm development following pollinations of diploids by tetraploids, and a much shortened proliferation phase and earlier ‘maturation’ of seeds resulting from maternal genomic excess (Scott et al., 1998). This difference between reciprocal pollinations is believed to result from many of the genes involved in early seed development being under maternal control, largely through gametic imprinting (Kermicle, 1970; Gutierrez-Marcos et al., 2003, 2006a; Costa et al., 2004; Gehring et al., 2004). The observations in Arabidopsis are in accordance with the kinship (or parental conflict) theory (Haig and Westoby, 1991; Haig, 2000), which holds that male and female gametes have different evolutionary ‘interests’, resulting in the differential epigenetic modification of male and female alleles of genes involved in resource acquisition from the maternal plant.
As the products of interploidy crosses in maize abort, these experiments provide little prima facie evidence pointing to maternal control of seed development, or in support of the kinship theory. However, as in Arabidopsis, the early development of these seeds differs depending on whether genomic excess is paternal or maternal. Measures of seed mass clearly show that following PGE crosses, mass of the seed increases significantly more rapidly than that of MGE pollinations, recalling the effects of imprinting seen in Arabidopsis (Scott et al., 1998). A number of genes involved in the early development of maize endosperms have been reported to be imprinted (Danilevskaya et al., 2003; Gutierrez-Marcos et al., 2003, 2004, 2006a; Haun et al., 2007) and this differential early seed growth in maize is likely to be regulated by the same mechanism. Why seeds from interploidy crosses from maize abort and those from Arabidopsis do not is unclear, but could be related to the transient nature of the Arabidopsis endosperm, compared with the persistent endosperm of maize and other cereal crop species. Thus, Arabidopsis, in which the endosperm appears to form a smaller component of the seed, may be able to survive a higher level of endosperm disruption than maize. In fact, Arabidopsis uniparental endosperms of maternal origin have recently been shown to be viable (Nowack et al., 2007).
Paternal and maternal genomic excess results in differential development of endosperm tissue domains
When the pVp1::GUS reporter line was used to pollinate tetraploid females the kernel developed extremely slowly, but the distribution of both the aleurone and the embryo (as reported by GUS activity) appeared normal, with the exception that the BETL showed significant levels of signal. BETL-specific cell-type formation is clearly inhibited in this region, and cells that would otherwise be BETL-specific assume an aleurone identity, confirming than an aleurone fate is the default condition for cells at the endosperm periphery (Costa et al., 2003; Gruis et al., 2006). This aspect of the effect of maternal genomic excess on BETL development was confirmed by expression of the pBETL1::GUS transgene being restricted to a small germinal domain of the highly reduced kernel.
Kernel development was also reduced following PGE pollinations and endosperms formed adopted an unusual gourd-shape between 6 and 10 DAP. While the pVp1::GUS transgene was expressed at the endosperm periphery, the GUS signal diffused into central endosperm, suggesting that cells in this region differ from those formed following other pollinations. In later stages (10–25 DAP) these endosperms became highly disorganized, as evidenced by the development of an irregular, fissured morphology, with the pVp1::GUS signal expressed at the surface. This breakdown in organization was also reflected by the pBETL1::GUS transgene expression, where signal was present not only in an irregular BETL, but also in ectopic domains throughout the endosperm, and at the distal tip.
The present study data support the data from Arabidopsis in which cellularization of the endosperm is delayed by an extended phase of nuclear proliferation following PGE crosses (Scott et al., 1998), and MGE results in endosperms that cellularize and form starch early. In maize, the lack of starch formation and diffusion of the GUS product into the central endosperm following PGE pollinations indicates an extended cellular proliferation phase, while the ‘normal’ (but very small) aspect of MGE endosperms at early stages, coupled with high levels of starch after only 10 DAP, points to early cellular differentiation. The second trend, which is not seen in Arabidopsis, relates to PGE endosperms where late development becomes strikingly irregular, resulting in ectopic expression of both the pVp1::GUS and the pBETL1::GUS transgenes, and the formation of fissures in the endosperm surface.
Endosperm-specific gene expression following interploidy crosses
The semi-quantitative RT-PCR generally confirmed the structural data. Comparison between endosperms resulting from within-ploidy crosses were generally consistent, with some evidence that ploidy may result in higher expression of genes such as BETL1, and perhaps ZmRP1, in the tetraploid.
Four days after MGE pollinations, the endosperms expressed only ZmESR1, but by 4–8 DAP expression of ZmMRP1, BETL1, BETL2 and ZmEND1 had commenced, but generally at lower levels than in either the diploid or the tetraploid controls. Interestingly, ZmESR1, despite expressing strongly at 4 DAP, becomes hardly detectable at 8 DAP, which may reflect aberrant development of the embryo, as embryoless kernels do not express ESR genes (Opsahl-Ferstad et al., 1997). By 10 DAP only small traces of BETL1 and ZmEND1 expression can be detected, suggesting that although the developing endosperm may appear normal, if reduced in size, the timing of the expression of a number of endosperm-specific genes is strongly affected by MGE. Current models proposed to explain cell fate specification in the endosperm suggest that the timing of events is critical, and that altered phasing of gene expression would result in aberrant development (Costa et al., 2003; Gutierrez-Marcos et al., 2006b).
Following PGE crosses, levels of expression of many of the genes tested (with the exception of END1) were higher than those of endosperms from diploid, and sometimes tetraploid, balanced crosses. The expression of BETL1 and 2 confirms the strong signals seen from the pBETL1::GUS transgene, and unsurprisingly the BETL-specific transcription factor ZmMRP1, which is held to activate genes in this domain (Gomez et al., 2002), is highly expressed after 6 d in these endosperms. Interestingly, ZmESR1, which defines a domain between the endosperm and embryo, continued to be highly expressed at 10 DAP, while its levels were falling following diploid and tetraploid balanced crosses. If events in maize parallel those in Arabidopsis and these endosperms undergo extended phases of cell proliferation (Scott et al., 1998), it is possible that ZmESR1 expression is a feature of this proliferative phase. Furthermore, as ESR genes are expressed only in the presence of developing embryos (Opsahl-Ferstad et al., 1997) continued ZmESR expression may signify further development of embryos than following MGE pollinations, which would be supported by the greater viability of PGE kernels.
An important caveat is that expression of a domain-specific marker sequence cannot be taken as evidence for function of a particular cell type. Indeed, our cytological work revealed that expression of both the reporter transgenes often preceded the development of characteristic and presumably functional cell structure.
The influence of maternal and paternal genomic excess on endosperm development
The present data collectively support the view that the early development of maize endosperms following interploidy crosses is regulated by the same principles as operate in Arabidopsis (Scott et al., 1998). However, the apparently unregulated morphological development of endosperms and the ectopic expression of the two reporter constructs following PGE crosses is not seen in Arabidopsis. In his early investigation of interploidy crosses, Cooper (1951) recorded the reduction of the BETL in maternal genomic excess kernels but provided no detailed information on the consequences of PGE pollinations. Likewise, Charlton et al. (1995) described detailed differences in BETL development following PGE crosses but did not record events elsewhere in the endosperm in similar detail. Recently, the ectopic expression of both ZmMEG1 and ZmMRP1 transcripts have been reported in PGE endosperms of maize using in situ hybridization (Gutierrez-Marcos et al., 2003, 2006b).
As balanced pollinations gave essentially similar outcomes, and comparison between diploid and tetraploid balanced crosses revealed that increased ploidy played a relatively minor role in regulating expression levels of the genes studied, some of the effects recorded may well be epigenetic in origin (Grant-Downton and Dickinson, 2005, 2006). The bulk of the differences observed between reciprocal interploidy pollinations during early development are likely to result from genomic imprinting (Scott et al., 1998; Alleman and Doctor, 2000; Gehring et al., 2004). Imprinting may also be responsible for the eventual abortion of MGE and PGE kernels for reasons discussed earlier, although uniparental maternal endosperms of Arabidopsis can be induced to bypass imprinting to form small but viable seeds (Nowack et al., 2007). However, even a combination of imprinting and dosage cannot explain the aberrant later development of PGE endosperms. A third factor has been proposed as controlling the outcomes of fertilization, i.e. the balance between maternal and paternal elements that regulate chromatin structure and function (Dilkes and Comai, 2004; Madlung and Comai, 2004), and which must differ significantly between plants, depending on the ‘genetic distance’ separating them. This balance is likely to be seriously disrupted following MGE and PGE. Certainly transcription factors are misregulated in interploidy crosses, as indicated by the ZmMRP1 expression data, and this will have important downstream effects [in the case of ZmMRP1, on the BETL sequences, (Gomez et al., 2002)]. Also, maternal genomes of the central cell are likely to possess a more comprehensive complement of these factors than that donated by the sperm (Engel et al., 2003). Thus, following a PGE pollination the level of these elements may become so attenuated as to be ineffective in regulating endosperm gene expression. Regulation of this type would involve both gene silencing and activation suppression, and absence of control at this level could be responsible for the irregular development of these endosperms and the ectopic expression of transcripts within them.
ACKNOWLEDGEMENTS
We thank BBSRC for financial support and the award of a Research Studentship to P.D.P., and Qing Zhang and Caroline O'Brien for technical assistance.
LITERATURE CITED
Author notes
Current address: HRI Warwick, Wellesbourne, Warwickshire CV35 9EF, UK.
Current address: NIAB, Huntingdon Road, Cambridge CB3 0LE, UK.