-
PDF
- Split View
-
Views
-
Cite
Cite
Sydney Brenner, In the Beginning Was the Worm …, Genetics, Volume 182, Issue 2, 1 June 2009, Pages 413–415, https://doi.org/10.1534/genetics.109.104976
Close - Share Icon Share
Abstract
In my end is my beginning.
T. S. Eliot
Anecdotal, Historical and Critical Commentaries on Genetics
CAENORHABDITIS elegans genetics started formally in the first few days of October 1967 with the first mutant hunt, which produced a grand total of two mutants. The first, E1, was named a “dumpy,” because of its distinctive body shape; the second, E2, was termed a “variable abnormal,” because of the range of defects found in its homozygous progeny. The low yield from this first mutant hunt, using EMS mutagenesis, simply showed how bad I was initially at discerning phenotypes, but I learned the art rather quickly and, on successive experiments, my yield rose to between 25 and 30 per screening. I learnt by doing and many others have since followed the same path and found that the understanding of wild-type behavior comes best after the discovery and analysis of mutations that alter it. I cannot really describe how triumphant I felt working with E1, backcrossing it to wild-type N2 with the male cultures I had previously established, and proving that the resulting heterozygotes segregated it in the classic Mendelian ratio of 1:3. Getting a mutant of a complex organism and confirming Gregor Mendel in only 2 weeks was most satisfying.
The genetics of diploids was novel to most of my molecular biologist contemporaries, who were accustomed only to the haploid genetics of phage and bacteria. Terms such as “dominant” and “recessive” soon became common currency in our lab. The self-fertilizing properties of the hermaphrodite, with the use of males to transport genes from one hermaphrodite to another, made genetic manipulation simple. When John Sulston discovered a method of freezing C. elegans, we could embark on serious genetic research. Actually what he discovered was a method of thawing frozen nematodes and preserving viability; everything survives freezing—it's the thawing that typically does the damage.
Over the next few years, I found many mutants, both directly and by special screens, and I developed methods to complement and map them. In 1974, I published an article entitled “The genetics of Caenorhabditis elegans” in this journal (Brenner 1974). It reported a study of 300 EMS-induced mutants and a map of about 100 genes on six linkage groups. It had two other interesting features. First, it unapologetically gave a full account of the methods that were used in hermaphrodite genetics, which today would be consigned to a remote database as supporting information. This has ensured that the article is cited, almost ritually, in almost every article written on C. elegans genetics, and this has given a 35-year-old article an unexpectedly long life. There used to be a similar source in bacteriophage genetics, published by Mark Adams in Methods in Medical Research. Since the reference was often copied from the reference list of current articles, it sometimes underwent mutation, and the change generated novel citation lineages that could be followed in the subsequent literature. I have not checked whether this has happened to Brenner (1974), but since nobody these days copies references by longhand and has them transcribed by a typist, I would not expect it in the era of word processing machines. The second interesting feature of this article is that, in the Introduction, it outlined how a reductionist program of research might be pursued in complex, multicellular organisms—or, at any rate, how we were going to pursue it for the C. elegans nervous system. It also proclaimed the now well-known advantages of C. elegans for the projected research, namely that it is small, rapidly growing, and easily handled in the laboratory. Today, all of this would be deleted by an editor as obvious, and yet I feel that it is important now, as I felt it was then, to give some indication as to why I was going to all the trouble of developing the genetics of a new model organism.
The Genetics article was accompanied in the journal by another article by John Sulston and myself titled “The DNA of Caenorhabditis elegans” (Sulston and Brenner 1974). Although strictly speaking, such work was not considered to be genetics at the time, the editor, Arthur Chovnick, agreed with us that the two should go together. The DNA article showed that the unique sequence fraction, some 83% of the DNA, was only 20 times the amount in Escherichia coli. We had no real idea then how large eukaryotic genes were, yet in the discussion of the Genetics article there is an argument that the genome of C. elegans could not be constituted of E. coli-sized genes of 1 kb or could be, as they were referred to at the time, NMBGs—naive molecular biologist's genes. I suggested that there must be a lot of noncoding DNA that was not susceptible to mutation or, at least, to mutations that would give phenotypes. I remember that I once compared the frequency of EMS-induced mutations in a myosin gene (unc-54) in C. elegans with that in the β-galactosidase gene of the accompanying bacteria and found that they were about the same. Since the proteins are of similar sizes, this showed that the target for mutation was the same and the organisms were essentially transparent to the mutagen. At that time, I was very concerned with the C-value paradox, the idea that the apparent complexity of organisms could not be correlated with the size of their genomes and that many organisms seemed to have more DNA than they reasonably needed. Physicists, in contrast, were concerned that organisms did not have enough DNA to explain their complexity but the clever molecular biologist, not to mention the thoughtful evolutionary biologist, knew that each organism had exactly the right amount for its needs.
What, the reader may ask, did we do in the 6 years between starting C. elegans genetics and publishing the first article on it? Since the animal has a short life cycle of 3.5 days, it should not have taken all that much time just to complement and map the mutations. Many visitors who came to the MRC Lab in Cambridge thought that we spent far too much time eating, drinking, and talking. Observing us only during normal working hours, you could see their point. If one arrived at the lab at the reasonable hour of 10 am, there was just time to open one's mail before adjourning to the canteen for morning coffee, usually prolonged by a very interesting discussion on some aspect of science. This did not leave much time before lunch, which naturally was also accompanied by discussion that was terminated only by rushing off to attend an afternoon seminar on the Bohr effect in hemoglobin or the like. That brought one to afternoon tea and after that there was hardly enough time to start anything in the lab before adjourning to the pub for liquid and intellectual refreshment. It was only after dinner that the real work started and the lab then filled up with the owls. Even these bouts of work had to be interrupted, of course, for midnight coffee and more discussions. Often, the few larks like myself, who came to lab very early in the morning, met the owls going home, and there were many nights when I became an owl as well.
The right answer is that we were very busy with many other aspects of the project at the same time. I could easily do my genetics in the morning, with my assistant Muriel Wigby, and I then spent a lot of my time getting the electron microscopy going with Nichol Thomson. When we looked at the mutants using polarized light microscopy, we quickly discovered that some of the paralyzed mutants had muscle defects and that this reflected effects of the mutations on the thick filaments, as was soon confirmed by electron microscopy. We began a long series of experiments on developing molecular biology in the nematode through the protein chemistry of myosin and other muscle components. Sandy McCleod was an early colleague, and Bob Waterston and Henry Epstein entered C. elegans research this way. This work culminated later in the cloning of the myosin gene by Jon Karn.
I also spent an inordinate amount of time with computers. We thought we would try to automate the reconstruction of serial section electron micrographs, for comparing mutant and wild-type nervous systems, and John White joined me for this purpose. He started by writing programs in binary for a Ferranti computer that Ulli Arndt was using to automate crystallography and that had originally seen service in Royal Navy submarines. I learnt computing and managed to persuade the MRC to buy a Modular I computer for us, which we were later able to expand very cheaply by talking to the “liquidator” (who had nothing to do with the present Governor of the State of California, but was a gentleman disposing of the assets of our now bankrupt computer company). I recklessly took on projects, writing in assembly language, such as altering Fortran compilers to interface with a graphics system that John White had implemented. David Marr joined the lab and Graeme Mitchison became a user, and we all spent many days and nights crawling on the floor, gluing pieces of paper tape together. Although I learned a lot, all of this turned out to be ridiculously premature and the reconstruction was finally done by hand, by Eileen Southgate and Rita Fishpool. Only now, with cheap computing power, is automatic reconstruction becoming feasible, and I am told that a “wiring diagram” of the C. elegans nervous system can be assembled in a week.
During this period, most people viewed C. elegans almost as a joke organism and, adding insult to insult, often confused it with the flatworm. Jim Watson, in particular, said he would not give me a penny for this work, complaining that I was 20 years ahead of my time. What changed everything was the advent of cloning and sequencing. It opened up all of genetics and gave us direct access to genes. We were no longer subject to the tyranny of the reproductive cycles of organisms, but could study the genomes of everything. In addition, an increasing number of talented and enthusiastic postdoctoral fellows came to join the lab and the next decade saw an explosive growth in the number of people in the field. Some years ago I was shown a lineage of C. elegans workers by Philip Anderson: Apart from the Cambridge group of Tony Stretton, John White, and John Sulston, and later Jonathan Hodgkin, the F1's included David Hirsh, Richard Russell, David Baillie, Don Riddle, Sam Ward, Donna Albertson, and Bob Waterston and later Bob Horvitz, Jon Karn, Barbara Meyer, Cynthia Kenyon, and many others. Some of these lineages extended to F4's, and they must be deeper now. (Perhaps if the compilers of this Almanac de Gotha of C. elegans workers are still pursuing this research, they could send me a copy.) I was told that there are now 400 C. elegans labs and that new ones are being established at a rate of one a week, more than McDonald's is achieving with new restaurants. I do not believe the last statement but would like to believe the first.
People often ask me why I left C. elegans research just when it was getting really interesting. The answer is simple: The people doing it were much better than I was, and attendance at one meeting showed me that their students were even better than they were. I have often quoted J. D. Bernal, who compared science to a game of chess: The middle game is extremely boring and it is given to very few of us to play the end game. There remains only the opening game, which, for me, is the most exciting part of scientific research. I think I am quite good at it, but, of course, it is interesting and satisfying only when it has a successful outcome. You have only to look at the articles in this and other journals to gauge what C. elegans researchers have achieved in changing biology, and it pleases me to know that they have succeeded in this not only through their own individual endeavors but also by creating a unique community of sharing scientists.
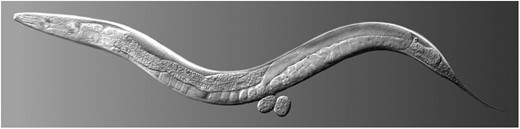
Wild type hermaphrodite Caenorhabditis elegans. Courtesy of Maria Gallegos.
References
Brenner, S.,
Sulston, J. E., and S. Brenner,