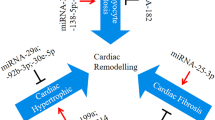
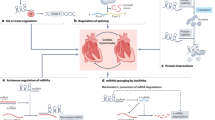
Abstract—
Hypertrophic cardiomyopathy (HCM) is the most common genetically determined heart pathology and is often accompanied by fatal complications. Today, the traditional view of the monogenic origin of HCM is being replaced by the idea of it as an oligogenic disease, the clinical phenotype of which is determined not only by mutations in the genes encoding sarcomere proteins in cardiomyocytes, but also by the contribution of other genes (other sarcomeric genes, non-sarcomeric protein-coding modifier genes, and regulatory non-coding RNA genes). Transcriptome analysis is an informative approach for elucidating the nature of HCM, which allows one to evaluate the expression of all genes, evaluate the effect of mutations in a gene on its transcript level, and reveal the mechanisms involved in the regulation of gene expression. This review presents an analysis of published data on the spectra of genes whose differential expression has been detected in the myocardium during the development of HCM in humans and model animals. Special attention is paid to the genes of non-coding regulatory RNAs: miRNAs and long non-coding RNAs, which may be involved in the pathogenesis of the disease. We analyzed studies devoted to the investigation of miRNA levels in the blood of HCM patients to explore the available diagnostic and prognostic biomarkers of the disease. The totality of the reviewed data, despite their relative scarcity, indicates the effectiveness of transcriptome profiling in studying the molecular mechanisms of HCM pathogenesis.

Similar content being viewed by others
REFERENCES
Elliott P.M., Anastasakis A., Borger M.A., Borggrefe M., Cecchi F., Charron P., Hagege A.A., Lafont A., Limongelli G., Mahrholdt H., McKenna W.J, Mogensen J., Nihoyannopoulos P., Nistri S., Pieper P.G., et al. 2014. ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: The task force for the diagnosis and management of hypertrophic cardiomyopathy of the European Society of Cardiology (ESC). Eur. Heart J. 35, 2733–2779.
Maron B.J., Gardin J.M., Flack J.M., Gidding S.S., Kurosaki T.T., Bild D.E. 1995. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4 111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 92, 785–789.
Semsarian C., Ingles J., Maron M.S., Maron B.J. 2015. New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 65, 1249–1254.
Charron P., Elliott P.M., Gimeno J.R., Caforio A.L.P., Kaski J.P., Tavazzi L., Tendera M., Maupain C., Laroche C., Rubis P., Jurcut R., Calo L., Helio T.M., Sinagra G., Zdravkovic M., et al. 2018. The Cardiomyopathy Registry of the EURObservational Research Programme of the European Society of Cardiology: Baseline data and contemporary management of adult patients with cardiomyopathies. Eur. Heart J. 39, 1784–1793.
Maron B.J., Doerer J.J., Haas T.S., Tierney D.M., Mueller F.O. 2009. Sudden deaths in young competitive athletes: analysis of 1 866 deaths in the United States, 1980‒2006. Circulation. 119, 1085–1092.
Lorenzini M., Anastasiou Z., O’Mahony C., Guttman O.P., Gimeno J.R., Monserrat L., Anastasakis A., Rapezzi C., Biagini E., Garcia-Pavia P., Limongelli G., Pavlou M., Elliott P.M., Hypertrophic Cardiomyopathy Outcomes investigators. 2019. Mortality among referral patients with hypertrophic cardiomyopathy vs. the general European population. JAMA Cardiol. e194534.
Raghow R. 2016. An ‘Omics’ perspective on cardiomyopathies and heart failure. Trends Mol. Med. 22, 813‒827.
Jarcho J.A., McKenna W., Pare J.A., Solomon S.D., Holcombe R.F., Dickie S., Levi T., Donis-Keller H., Seidman J.G., Seidman C.E. 1989. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N. Engl. J. Med. 321, 1372–1378.
Geisterfer-Lowrance A.A., Kass S., Tanigawa G., Vosberg H.P., McKenna W., Seidman C.E., Seidman J.G. 1990. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 62, 999–1006.
Solomon S.D., Jarcho J.A., McKenna W., Geisterfer-Lowrance A., Germain R., Salerni R., Seidman J.G., Seidman C.E. 1990. Familial hypertrophic cardiomyopathy is a genetically heterogeneous disease. J. Clin. Invest. 86, 993–999.
Seidman C.E., Seidman J.G. 2011. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: A personal history. Circ. Res. 108, 743–750.
Lopes L.R., Rahman M.S., Elliott P.M. 2013. A systematic review and meta-analysis of genotype-phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart. 99, 1800–1811.
Wolf C.M. 2019. Hypertrophic cardiomyopathy: Genetics and clinical perspectives. Cardiovasc. Diagn. Ther. 9, S388–S415.
Palmer B.M., Wang Y., Teekakirikul P., Hinson J.T., Fatkin D., Strouse S., Vanburen P., Seidman C.E., Seidman J.G., Maughan D.W. 2008. Myofilament mechanical performance is enhanced by R403Q myosin in mouse myocardium independent of sex. Am. J. Physiol. Heart Circ. Physiol. 294, H1939–H1947.
Green E.M., Wakimoto H., Anderson R.L., Evanchik M.J., Gorham J.M., Harrison B.C., Henze M., Kawas R., Oslob J.D., Rodriguez H.M., Song Y., Wan W., Leinwand L.A., Spudich J.A., McDowell R.S., et al. 2016. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 351, 617–621.
Tyska M.J., Hayes E., Giewat M., Seidman C.E., Seidman J.G., Warshaw D.M. 2000. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ. Res. 86, 737–744.
Tardiff J.C. 2005. Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes. Heart Fail. Rev. 10, 237–248.
Sequeira V., Wijnker P.J., Nijenkamp L.L., Kuster D.W., Najafi A., Witjas-Paalberends E.R., Regan J.A., Boontje N., Ten Cate F.J., Germans T., Carrier L., Sadayappan S., van Slegtenhorst M.A., Zaremba R., Foster D.B., et al. 2013. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ. Res. 112, 1491–1505.
Linke W.A. 2008. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc. Res. 77, 637–648.
Fatkin D., McConnell B.K., Mudd J.O., Semsarian C., Moskowitz I.G., Schoen F.J., Giewat M., Seidman C.E., Seidman J.G. 2000. An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J. Clin. Invest. 106, 1351–1359.
Crilley J.G., Boehm E.A., Blair E., Rajagopalan B., Blamire A.M., Styles P., McKenna W.J., Ostman-Smith I., Clarke K., Watkins H. 2003. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J. Am. Coll. Cardiol. 41, 1776–1782.
Ashrafian H., McKenna W.J., Watkins H. 2011. Disease pathways and novel therapeutic targets in hypertrophic cardiomyopathy. Circ. Res. 109, 86–96.
Ferrantini C., Belus A., Piroddi N., Scellini B., Tesi C., Poggesi C. 2009. Mechanical and energetic consequences of HCM-causing mutations. J. Cardiovasc. Transl. Res. 2, 441–451.
Marston S.B. 2011. How do mutations in contractile proteins cause the primary familial cardiomyopathies? J. Cardiovasc. Transl. Res. 4, 245–255.
Walsh R., Thomson K.L., Ware J.S., Funke B.H., Woodley J., McGuire K.J., Mazzarotto F., Blair E., Seller A., Taylor J.C., Minikel E.V., MacArthur D.G., Farrall M., Cook S.A., Watkins H. 2017. Reassessment of Mendelian gene pathogenicity using 7855 cardiomyopathy cases and 60 706 reference samples. Genet. Med. 19, 192–203.
Van Driest S.L., Ackerman M.J., Ommen S.R., Shakur R., Will M.L., Nishimura R.A., Tajik A.J., Gersh B.J. 2002. Prevalence and severity of “benign” mutations in the beta-myosin heavy chain, cardiac troponin T, and alpha-tropomyosin genes in hypertrophic cardiomyopathy. Circulation. 106, 3085–3090.
Oliva-Sandoval M.J., Ruiz-Espejo F., Monserrat L., Hermida-Prieto M., Sabater M., Garcia-Molina E., Ortiz M., Rodriguez-Garcia M.I., Nunez L., Gimeno J.R., Castro-Beiras A., Valdes M. 2010. Insights into genotype-phenotype correlation in hypertrophic cardiomyopathy. Findings from 18 Spanish families with a single mutation in MYBPC3. Heart. 96, 1980–1984.
Jansweijer J.A., van Spaendonck-Zwarts K.Y., Tanck M.W.T., van Tintelen J.P., Christiaans I., van der Smagt J., Vermeer A., Bos J.M., Moss A.J., Swan H., Priori S.G., Rydberg A., Tfelt-Hansen J., Ackerman M.J., Olivotto I., et al. 2019. Heritability in genetic heart disease: The role of genetic background. Open Heart. 6, e000929.
Ingles J., Burns C., Bagnall R.D., Lam L., Yeates L., Sarina T., Puranik R., Briffa T., Atherton J.J., Driscoll T., Semsarian C. 2017. Nonfamilial hypertrophic cardiomyopathy: Prevalence, natural history, and clinical implications. Circ. Cardiovasc. Genet. 10.
Bagnall R.D., Ingles J., Dinger M.E., Cowley M.J., Ross S.B., Minoche A.E., Lal S., Turner C., Colley A., Rajagopalan S., Berman Y., Ronan A., Fatkin D., Semsarian C. 2018. Whole genome sequencing improves outcomes of genetic testing in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 72, 419–429.
Maron B.J., Maron M.S., Maron B.A., Loscalzo J. 2019. Moving beyond the sarcomere to explain heterogeneity in hypertrophic cardiomyopathy: JACC review topic of the week. J. Am. Coll. Cardiol. 73, 1978–1986.
Pasipoularides A. 2018. Challenges and controversies in hypertrophic cardiomyopathy: Clinical, genomic and basic science perspectives. Rev. Esp. Cardiol. Engl. Ed. 71, 132–138.
Burns C., Bagnall R.D., Lam L., Semsarian C., Ingles J. 2017. Multiple gene variants in hypertrophic cardiomyopathy in the era of next-generation sequencing. Circ. Cardiovasc. Genet. 10, S307.
Mouton J.M., van der Merwe L., Goosen A., Revera M., Brink P.A., Moolman-Smook J.C., Kinnear C. 2016. MYBPH acts as modifier of cardiac hypertrophy in hypertrophic cardiomyopathy (HCM) patients. Hum. Genet. 135, 477–483.
Wooten E.C., Hebl V.B., Wolf M.J., Greytak S.R., Orr N.M., Draper I., Calvino J.E., Kapur N.K., Maron M.S., Kullo I.J., Ommen S.R., Bos J.M., Ackerman M.J., Huggins G.S. 2013. Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 6, 10–18.
Perkins M.J., Van Driest S.L., Ellsworth E.G., Will M.L., Gersh B.J., Ommen S.R., Ackerman M.J. 2005. Gene-specific modifying effects of pro-LVH polymorphisms involving the renin-angiotensin-aldosterone system among 389 unrelated patients with hypertrophic cardiomyopathy. Eur. Heart J. 26, 2457–2462.
Ortlepp J.R., Vosberg H.P., Reith S., Ohme F., Mahon N.G., Schroder D., Klues H.G., Hanrath P., McKenna W.J. 2002. Genetic polymorphisms in the renin-angiotensin-aldosterone system associated with expression of left ventricular hypertrophy in hypertrophic cardiomyopathy: A study of five polymorphic genes in a family with a disease causing mutation in the myosin binding protein C gene. Heart. 87, 270–275.
Kolder I.C., Michels M., Christiaans I., Ten Cate F.J., Majoor-Krakauer D., Danser A.H., Lekanne Deprez R.H., Tanck M., Wilde A.A., Bezzina C.R., Dooijes D. 2012. The role of renin-angiotensin-aldosterone system polymorphisms in phenotypic expression of MYBPC3-related hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 20, 1071–1077.
Helms A.S., Day S.M. 2016. Hypertrophic cardiomyopathy: Single gene disease or complex trait? Eur. Heart J. 37, 1823–1825.
Cerrone M., Remme C.A., Tadros R., Bezzina C.R., Delmar M. 2019. Beyond the one gene-one disease paradigm: Complex genetics and pleiotropy in inheritable cardiac disorders. Circulation. 140, 595–610.
Consortium E.P. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74.
Costa F.F. 2010. Non-coding RNAs: Meet thy masters. Bioessays. 32, 599‒608.
Brosnan C.A., Voinnet O. 2009. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 21, 416–425.
Bos J.M., Hebl V.B., Oberg A.L., Sun Z., Herman D.S., Teekakirikul P., Seidman J.G., Seidman C.E., Dos Remedios C.G., Maleszewski J.J., Schaff H.V., Dearani J.A., Noseworthy P.A., Friedman P.A., Ommen S.R., et al. 2020. Marked up-regulation of ACE2 in hearts of patients with obstructive hypertrophic cardiomyopathy: Implications for SARS-CoV-2-mediated COVID-19. Mayo Clin. Proc. 95, 1354–1368.
Malgija B., Kumar N.S., Piramanayagam S. 2018. Collective transcriptomic deregulation of hypertrophic and dilated cardiomyopathy: Importance of fibrotic mechanism in heart failure. Comput. Biol. Chem. 73, 85–94.
Ren C.W., Liu J.J., Li J.H., Li J.W., Dai J., Lai Y.Q. 2016. RNAseq profiling of mRNA associated with hypertrophic cardiomyopathy. Mol. Med. Rep. 14, 5573–5586.
Liu Y., Morley M., Brandimarto J., Hannenhalli S., Hu Y., Ashley E.A., Tang W.H., Moravec C.S., Margulies K.B., Cappola T.P., Li M., MAGNet consortium. 2015. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 105, 83–89.
Seeger T., Shrestha R., Lam C.K., Chen C., McKeithan W.L., Lau E., Wnorowski A., McMullen G., Greenhaw M., Lee J., Oikonomopoulos A., Lee S., Yang H., Mercola M., Wheeler M., et al. 2019. A premature termination codon mutation in MYBPC3 causes hypertrophic cardiomyopathy via chronic activation of nonsense-mediated decay. Circulation. 139, 799–811.
Han L., Li Y., Tchao J., Kaplan A.D., Lin B., Li Y., Mich-Basso J., Lis A., Hassan N., London B., Bett G.C., Tobita K., Rasmusson R.L., Yang L. 2014. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc. Res. 104, 258‒269.
Dieseldorff Jones K.M., Vied C., Valera I.C., Chase P.B., Parvatiyar M.S., Pinto J.R. 2020. Sexual dimorphism in cardiac transcriptome associated with a troponin C murine model of hypertrophic cardiomyopathy. Physiol. Rep. 8, e14396.
Axelsson A., Iversen K., Vejlstrup N., Ho C., Norsk J., Langhoff L., Ahtarovski K., Corell P., Havndrup O., Jensen M., Bundgaard H. 2015. Efficacy and safety of the angiotensin II receptor blocker losartan for hypertrophic cardiomyopathy: The INHERIT randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol. 3, 123–131.
Maron M.S., Chan R.H., Kapur N.K., Jaffe I.Z., McGraw A.P., Kerur R., Maron B.J., Udelson J.E. 2018. Effect of spironolactone on myocardial fibrosis and other clinical variables in patients with hypertrophic cardiomyopathy. Am. J. Med. 131, 837–841.
Vakrou S., Fukunaga R., Foster D.B., Sorensen L., Liu Y., Guan Y., Woldemichael K., Pineda-Reyes R., Liu T., Tardiff J.C., Leinwand L.A., Tocchetti C.G., Abraham T.P., O’Rourke B., Aon M.A., Abraham M.R. 2018. Allele-specific differences in transcriptome, miRNome, and mitochondrial function in two hypertrophic cardiomyopathy mouse models. JCI Insight. 3, e94493
Sasagawa S., Nishimura Y., Okabe S., Murakami S., Ashikawa Y., Yuge M., Kawaguchi K., Kawase R., Okamoto R., Ito M., Tanaka T. 2016. Downregulation of gstk1 is a common mechanism underlying hypertrophic cardiomyopathy. Front. Pharmacol. 7, 162.
Zhao Y., Wang Z., Zhang W., Zhang L. 2019. MicroRNAs play an essential role in autophagy regulation in various disease phenotypes. Biofactors. 45, 844–856.
Kuster D.W., Mulders J., Ten Cate F.J., Michels M., Dos Remedios C.G., da Costa Martins P.A., van der Velden J., Oudejans C.B. 2013. MicroRNA transcriptome profiling in cardiac tissue of hypertrophic cardiomyopathy patients with MYBPC3 mutations. J. Mol. Cell. Cardiol. 65, 59–66.
Song L., Su M., Wang S., Zou Y., Wang X., Wang Y., Cui H., Zhao P., Hui R., Wang J. 2014. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J. Cell. Mol. Med. 18, 2266–2274.
Bagnall R.D., Tsoutsman T., Shephard R.E., Ritchie W., Semsarian C. 2012. Global microRNA profiling of the mouse ventricles during development of severe hypertrophic cardiomyopathy and heart failure. PLoS One. 7, e44744.
Palacin M., Reguero J.R., Martin M., Diaz Molina B., Moris C., Alvarez V., Coto E. 2011. Profile of microRNAs differentially produced in hearts from patients with hypertrophic cardiomyopathy and sarcomeric mutations. Clin. Chem. 57, 1614–1616.
Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D., Elia L., Latronico M.V., Hoydal M., Autore C., Russo M.A., et al. 2007. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618.
Gao J., Collyer J., Wang M., Sun F., Xu F. 2020. Genetic dissection of hypertrophic cardiomyopathy with myocardial RNA-Seq. Int. J. Mol. Sci. 21, e3040.
Roncarati R., Viviani Anselmi C., Losi M.A., Papa L., Cavarretta E., Da Costa Martins P., Contaldi C., Saccani Jotti G., Franzone A., Galastri L., Latronico M.V., Imbriaco M., Esposito G., De Windt L., et al. 2014. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 63, 920–927.
Derda A.A., Thum S., Lorenzen J.M., Bavendiek U., Heineke J., Keyser B., Stuhrmann M., Givens R.C., Kennel P.J., Schulze P.C., Widder J.D., Bauersachs J., Thum T. 2015. Blood-based microRNA signatures differentiate various forms of cardiac hypertrophy. Int. J. Cardiol. 196, 115–122.
Fang L., Ellims A.H., Moore X.L., White D.A., Taylor A.J., Chin-Dusting J., Dart A.M. 2015. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J. Transl. Med. 13, 314.
Yao R.W., Wang Y., Chen L.L. 2019. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551.
Kitow J., Derda A.A., Beermann J., Kumarswarmy R., Pfanne A., Fendrich J., Lorenzen J.M., Xiao K., Bavendiek U., Bauersachs J., Thum T. 2016. Mitochondrial long noncoding RNAs as blood based biomarkers for cardiac remodeling in patients with hypertrophic cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 311, H707–H712.
Yang W., Li Y., He F., et al. 2015. Microarray profiling of long non-coding RNA (lncRNA) associated with hypertrophic cardiomyopathy. BMC Cardiovasc. Disord. 15, 62.
Liu X., Ma Y., Yin K., Li W., Chen W., Zhang Y., Zhu C., Li T., Han B., Liu X., Wang S., Zhou Z. 2019. Long non-coding and coding RNA profiling using strand-specific RNA-seq in human hypertrophic cardiomyopathy. Sci. Data. 6, 90.
Hu X., Shen G., Lu X., Ding G., Shen L. 2019. Identification of key proteins and lncRNAs in hypertrophic cardiomyopathy by integrated network analysis. Arch. Med. Sci. 15, 484–497.
Sonnenschein K., Wilczek A.L., de Gonzalo-Calvo D., Pfanne A., Derda A.A., Zwadlo C., Bavendiek U., Bauersachs J., Fiedler J., Thum T. 2019. Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci. Rep. 9, 20350.
Li J., Wu Z., Zheng D., Sun Y., Wang S., Yan Y. 2019. Bioinformatics analysis of the regulatory lncRNA–miRNA–mRNA network and drug prediction in patients with hypertrophic cardiomyopathy. Mol. Med. Rep. 20, 549–558.
Shi H., Li J., Song Q., Cheng L., Sun H., Fan W., Li J., Wang Z., Zhang G. 2019. Systematic identification and analysis of dysregulated miRNA and transcription factor feed-forward loops in hypertrophic cardiomyopathy. J. Cell. Mol. Med. 23, 306–316.
Sun D., Li C., Liu J., Wang Z., Liu Y., Luo C., Chen Y., Wen S. 2019. Expression profile of microRNAs in hypertrophic cardiomyopathy and effects of microRNA-20 in inducing cardiomyocyte hypertrophy through regulating gene MFN2. DNA Cell Biol. 38, 796–807.
Funding
This study was supported by the Russian Science Foundation (grant no. 20-15-00353).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Transalted by M. Novikova
Abbreviations: SCD, sudden cardiac death; HCM, hypertrophic cardiomyopathy; iPSCs, induced pluripotent stem cells; LV, left ventricle; RAAS, renin-angiotensin-aldosterone system; lncRNA, long non-coding RNA; ncRNA, non-coding RNA; NGS, next generation sequencing.
Rights and permissions
About this article
Cite this article
Baulina, N.M., Kiselev, I.S., Chumakova, O.S. et al. Hypertrophic Cardiomyopathy as an Oligogenic Disease: Transcriptomic Arguments. Mol Biol 54, 840–850 (2020). https://doi.org/10.1134/S0026893320060023
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893320060023