-
PDF
- Split View
-
Views
-
Cite
Cite
Dietrich Rebholz-Schuhmann, Harald Kirsch, Miguel Arregui, Sylvain Gaudan, Mark Riethoven, Peter Stoehr, EBIMed—text crunching to gather facts for proteins from Medline, Bioinformatics, Volume 23, Issue 2, January 2007, Pages e237–e244, https://doi.org/10.1093/bioinformatics/btl302
Close - Share Icon Share
Abstract
Summary: To allow efficient and systematic retrieval of statements from Medline we have developed EBIMed, a service that combines document retrieval with co-occurrence-based analysis of Medline abstracts. Upon keyword query, EBIMed retrieves the abstracts from EMBL-EBI's installation of Medline and filters for sentences that contain biomedical terminology maintained in public bioinformatics resources. The extracted sentences and terminology are used to generate an overview table on proteins, Gene Ontology (GO) annotations, drugs and species used in the same biological context. All terms in retrieved abstracts and extracted sentences are linked to their entries in biomedical databases. We assessed the quality of the identification of terms and relations in the retrieved sentences. More than 90% of the protein names found indeed represented a protein. According to the analysis of four protein–protein pairs from the Wnt pathway we estimated that 37% of the statements containing such a pair mentioned a meaningful interaction and clarified the interaction of Dkk with LRP. We conclude that EBIMed improves access to information where proteins and drugs are involved in the same biological process, e.g. statements with GO annotations of proteins, protein–protein interactions and effects of drugs on proteins.
Availability: Available at Author Webpage
Supplementary Data: Supplementary Data are available at Bioinformatics online.
Contact: Rebholz@ebi.ac.uk
1 INTRODUCTION
Ready access to the scientific literature in conjunction with reliable and fast document retrieval methods is crucial for efficient research work. For even better support of researchers, teams of curators propagate results and hypotheses from the literature into electronic databases and thus allow access to consistent and comprehensive data (Rebholz-Schuhmann et al., 2005). Altogether, both researchers and curators profit from advancements in text mining and from new services attached to the scientific literature.
PubMed1 [Author Webpage] is the central access point for almost all biomedical publications, and its host, the NLM, has developed an efficient interface for customized and fast queries. The result of such a query is either a list of abstract titles or another format provided from PubMed (e.g. the abstracts in their XML format). In the case of an appealing title, the user can display the abstract which was written by the author to state the most important findings of the publication and thus contains statements of the full paper in a condensed form. On the basis of this information the PubMed user takes the decision to retrieve and read the full paper. As a result the full potential of Medline abstracts not only lies in merely selecting interesting publications but also in further exploitation for the contained information. This observation induced the development of new solutions that process Medline abstracts to reduce the amount of data and to provide part of the meaningful information (Craven and Krumlien, 1999).
Solutions have been made available that rely purely on the identification of terms, e.g. co-occurrence of terms or representation of an abstract by keywords. Co-occurrence of two terms is their mention in the same context, which can be either the whole abstract or a single sentence or phrase. Co-occurrence has been applied to prove its advantage for information retrieval (Stapley and Benoit, 2000; Jelier et al., 2005).
The text-mining community has made many attempts to improve access to information for researchers, leading to the development of the text-mining solutions PubGene, iHOP, BioIE and PubMatrix, which are briefly described below for an overview on current online services.
PubGene identifies co-occurring gene names in the same abstract to suggest networks of related genes (Jenssen et al., 2001). Its evaluation is based on the percentage of correctly identified network links. Apart from gene–gene associations, other types of annotation for genes or proteins have been proposed to be advantageous (Andrade and Valencia, 1998; Rindflesch et al., 2000). None of these solutions are available in the latest versions of publicly available literature resources.
iHOP is a service that offers access to a database of sentences, each containing co-occurring protein names in conjunction with additional keywords that indicate an interaction between them (Hoffmann and Valencia, 2004). An evaluation is not available, which makes it difficult to compare. By the nature of this approach, the data has to be regenerated from Medline upon any of its new releases.
BioIE retrieves abstracts from PubMed based on a keyword query and then identifies and extracts sentences that match predefined language patterns (Divoli and Attwood, 2005). Owing to its non-conformity to comprehensive terminological resources in the biomedical field, such as the Gene Ontology (GO) or UniProtKB/Swiss-Prot, this solution generates results that are not well supported by the domain knowledge of molecular biology (Ashburner et al., 2000).
PubMatrix accepts terms from the user and then performs PubMed queries with all pairs of terms to finally build a matrix of counts referring to the co-occurrence of each pair (Becker et al., 2003). Selection of an entry in the matrix by the user starts the retrieval of the respective Medline abstracts. This approach generates a high reading workload.
Other solutions have been proposed that identify and extract pieces of information from an abstract. Such data can be fed into databases. Unfortunately, none of these solutions is publicly accessible (Gaizauskas et al., 2003; Friedman et al., 2001; Rzhetsky et al., 2004). In summary, no solution is available that efficiently combines document retrieval with information extraction to select relevant sentences from Medline and link the terminology in these sentences to information in the public biomedical databases. To fill this gap we developed a novel online service, EBIMed, which speeds up access to information from Medline.
Users of EBIMed apply keyword searches to retrieve a set of abstracts, which are then filtered for sentences containing UniProtKB/Swiss-Prot proteins, GO terms, drugs and species. All identified terms, sentences and abstracts are displayed in tables and all terms are linked to the entry in the biomedical database from which the term was derived. Altogether, users need not skim through lists of abstracts anymore.
2 DESIGN PRINCIPLES
The information contained in Medline abstracts is conveyed to a great part by biomedical terminology such as protein names and GO terms. It is EBIMed's goal to make this information accessible by extracting, ranking and organizing these key terms.
EBIMed labels UniProtKB/Swiss-Prot protein name in the text if it co-occurs with another UniProtKB/Swiss-Prot protein name, a GO term, a drug or species name. Such co-occurrences can be interpreted as two proteins being involved in the same biological process (e.g. protein–protein interactions), as functional annotations (GO annotations), as proteins being targeted by drugs (drug–protein relations) and as proteins of model organisms. Other types of terminology could be integrated. This would require better understanding of how these terms (e.g. diseases) are related to proteins and how well the terminology is supported.
The user can either submit a keyword query or a list of PubMed identifiers (PMIDs) to start the process. The query terms are not used as parameters for the analysis and therefore need not contain any protein name, drug, species or GO term. The only dependence between the query and EBIMed's analysis is the set of retrieved abstracts.
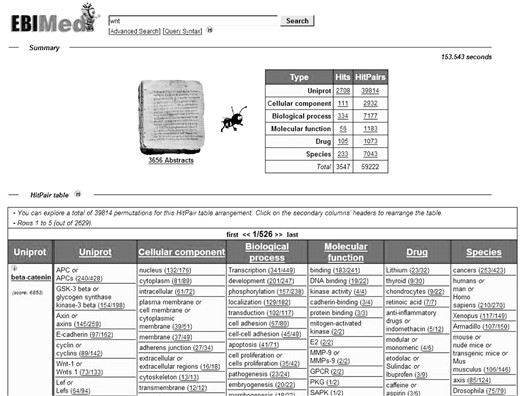
Terms that occur in the same sentence form a pair (co-occurrence). All sentences containing pairs are gathered, sorted and grouped according to the identified pairs in the sentence. Every pair of concepts is presented in a table and pairs with the highest number of evidence sentences are ranked highest and listed first (Fig. 1). For each pair in the table, a link is provided to a list of sentences containing the pair. Each sentence is linked to its original abstract.
In EBIMed the keyword query Wnt returns 4675 Medline abstracts. UniProtKB/Swiss-Prot proteins are extracted from all abstracts (3275 unique proteins) as well as all pairs of UniProtKB/Swiss-Prot proteins stated in a single sentence (53 649 unique pairs). In the table all entries in the columns 2–6 (e.g. ‘APC’ or ‘Lithium’) form a pair with ‘beta-catenin’ (column 1). The numbers behind the pairs indicate the number of sentences that contain this pair (right number) and the number of abstracts that contain the sentences (left number). The two numbers link to the list of sentences that again are linked to the full abstract. Abstracts and sentences are marked up with identified terminology and provide links to the terminological or database resource where the terms were taken from.
Initially, the leftmost column of the table lists a UniProtKB/Swiss-Prot protein and all other columns to the right list the co-occurring concepts (e.g. protein names, GO terms, drug names and species names) that form a pair with the protein. A column to the right will become the leading leftmost column as soon as the user selects it with a mouse click on the header of the column. The content of the table is then reorganized to match the concepts in the leftmost column. Above the table a display shows the total number of abstracts analysed and a list with the number of pairs encountered for the different types of terms.
In principle, the number of pairs increases with the number of retrieved abstracts and the number of identified terms from the domain of molecular biology. For example, the query Wnt currently retrieves 4675 abstracts (date of retrieval: 12 March 2005) with 3275 listed UniProtKB/Swiss-Prot proteins, RNAi leads to the selection of 2511 abstracts dealing with 2821 identified proteins, whereas the gynecological treatment ‘cerclage’ induces the retrieval of 1478 abstracts with only 80 UniProtKB/Swiss-Prot entries.
3 IMPLEMENTATION
Medline abstracts are provided by the NLM with periodic updates. The local Medline installation is indexed with Lucene (Hatcher and Gospodnetic, 2004). The index currently covers title, abstract text, author list, affiliation and MeSH terms. Tokenization as part of the indexing process normalizes tokens to lowercase and for terms described by the regular expression ‘[a-z]+[0-9]+’, such as gene and protein names, the tokenizer separates the character string from the digits. Retrieval of abstracts reporting on such terms is done by proximity search leading to the same retrieval result if either the query ‘HZF1’ or ‘HZF-1’ is applied. WordNet (Author Webpage) is used to normalize irregular verbs to their base form as well as plural forms to singular.
The retrieved abstracts are processed in a set of cascaded modules (Hopcroft and Ullman, 2001). They are set up in a pipeline and each of them reads and writes XML code (Kirsch et al., 2006). Identification of UniProtKB/Swiss-Prot proteins is based on the content of the UniProtKB/Swiss-Prot XML file, which provides the current set of protein names and synonyms. The names are generalized according to the following rules. White space characters are replaced by a selection of optional characters (‘ ’, ‘-’, ‘_’, ‘/’) leading, for example, to the regular expression ‘HZF[- _/]?1’ for ‘HZF-1’ and for ‘HZF 1’.
Identified protein names are marked up and are linked to the corresponding UniProtKB/Swiss-Prot database server entry (Author Webpage). In the case of ambiguous acronyms, e.g. ‘ESC’ for ‘embryonic stem cells’, the expanded form found in conjunction with the acronym is then used for disambiguation (Gaudan et al., 2005). Other ambiguous protein names that are stated without their expanded version, and those that have a high frequency in the British National Corpus (Author Webpage), are excluded as well, such as ‘BY’, ‘AND’ and others. For a complete description refer Rebholz-Schuhmann et al. (2006).
Identification of GO terms is based on matching of uppercase and lowercase GO-term variants, which currently meets the demands expressed by curators. In the case of multiple matches the leftmost longest match is chosen. Drug names are provided from MedlinePlus (Author Webpage). The NCBI taxonomy (Author Webpage) serves as terminological resource for the identification of species. The priority rules for the matching of the terminology are UniProtKB/Swiss-Prot first, then GO:cellular component, GO:biological process, GO:molecular function, drugs and species (Rebholz-Schuhmann et al., 2006).
EBIMed makes extensive use of lists of synonyms for single terms, such as protein names, and of categories of terms, for example drug names. The synonym lists fully follow the standards provided by the orginal source (see above). In its overview display (Fig. 1) EBIMed presents only those terms that have actually been encountered in the text and presents only a normalized version, e.g. only lowercase representations, whereas the corresponding sentence might contain a morphological variant of the displayed term.
EBIMed's main table is sorted from the top to the bottom according to the total number of pairs linked to the concept in the leftmost column. Initially this column contains UniProtKB/Swiss-Prot proteins and the protein with the largest number of extracted pairs is on the top of the table. All abstracts provided from EBIMed in a list are ranked according to the relevance of the abstracts to the initial query, i.e. according to their Lucene score (Hatcher and Gospodnetic, 2004). If several documents share the same score then they are sorted in inverse chronological order (newest document first). Listed sentences follow the order of their abstracts. Sentences from a single abstract are grouped together and sorted according to their order in the text.
For advanced queries EBIMed provides a special interface. In this interface the user can specify queries that are only directed towards the abstract title, the abstract text, the author list and the MeSH terms (similar to PubMed). Furthermore, the total number of retrieved abstracts can be raised from the default of 500 abstracts to 10 000 abstracts.
4 EVALUATION OF EBIMED: ASSESSMENT OF FACTS RETRIEVED FROM THE Wnt PATHWAY
EBIMed is evaluated against its goal of offering ready access to the information contained in abstracts by extracting, ranking and organizing biomedical terminology. In contrast to the evaluation of a retrieval engine, we do not measure the relevance of retrieved abstracts as compared to a desired result set, but judge the quality of the extracted information with regard to a biomedical topic. For the topic we chose the Wnt pathway and used the keyword query Wnt to retrieve abstracts with EBIMed. The assessment is based on four analyses described below that provide numbers to describe the precision2 and recall3 of terms and relations identified.
We chose the Wnt pathway as a test case for several reasons. The keyword describes a reasonably self-contained topic. The number of abstracts returned by the Wnt query (4675) is high enough to provide statistically meaningful counts.
From the description of the procedure below, it will become clear that the assessment concentrates on the relations between entities that are meaningful in biology and medicine, like proteins and drugs, because these relations are what EBIMed intends to summarize. For a completely unbiased analysis it would be necessary to select relations between, for example, proteins truly at random from an independent source that contains all relations from the literature and to then measure how well EBIMed recovers them from Medline. Unfortunately such a source does not exist, taking into consideration that public databases (e.g. IntACT) are biased towards their curation goals.
For the assessment it is necessary to understand that EBIMed's tables gather facts that have a heterogeneous distribution in Medline. In other words, some facts are described many times in the literature, under various forms, whereas others are mentioned only once or twice. EBIMed is designed to collect them all without preference. In this respect the query Wnt is well suited for our assessment, since facts in the Wnt pathway (e.g. protein–protein interactions) show a similar distribution: there is a range of publications from 1757 for protein beta-catenin to only 11 for protein PP2A in the context of Wnt. Therefore, we can be confident that a random selection of aspects from the results of the Wnt query provides for a valid assessment.
The Wnt pathway is described in the Kyoto Encyclopedia of Genes and Genomes [(KEGG) (Author Webpage, KEGG Release 37.0, January 2006)] as well as in the Signal Transduction Knowledge Environment [(STKE) (Author Webpage)], which allows for validation of retrieved facts with the representation in these public resources.
4.1 Assessment procedure: how?
For the assessment we manually evaluated results returned by EBIMed in four different analyses.
4.1.1 Analysis-1
Because EBIMed generates tables based on occurrences of terms in the abstracts, we first assessed how accurately EBIMed identifies terms. We queried using Wnt, selected abstracts containing not more than 200 sentences (191 sentences in 31 abstracts) according to their sorting from the complete list of abstracts (4675), and counted how many terms (proteins/genes, GO terms, drugs, species) were correctly identified or missed by EBIMed.
4.1.2 Analysis-2
In addition to single terms, EBIMed lists pairs of terms co-occurring in sentences as an assumed relation between both concepts. To measure the rate of true relations we again applied the same query Wnt, selected the first 20 proteins in alphabetical order identified by EBIMed and manually analysed all provided sentences that contain protein pairs (94 sentences). Note that our selection does not restrict the second protein in the pair to the list of 20 selected proteins.
4.1.3 Analysis-3
Similar to analysis-2 we estimated the precision of drug–protein relations identified by EBIMed. Again we applied the query Wnt. For all retrieved sentences containing a drug–protein pair (total 118) we manually checked for a meaningful relation.
4.1.4 Analysis-4
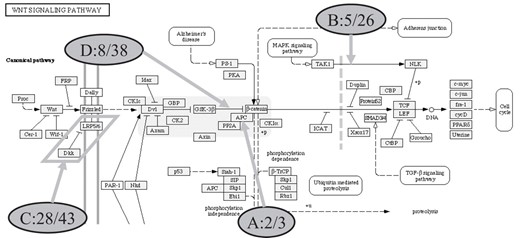
Finally, we assessed the retrieval of facts for documented protein–protein interactions from the Wnt pathway. As representatives we chose protein pairs that have been described in KEGG as well as in STKE, where a pair is either two nodes linked by an edge or two nodes side by side. We identified 108 unique pairs and 10 pairs common to both sources. We measured how many of these pairs were reproduced by EBIMed upon the query Wnt. In addition, we randomly selected 4 interaction pairs out of the 10 pairs confirmed in both sources (Fig. 2) and used both protein names in conjunction with Wnt for a combined query such as Wnt AND APC AND PP2A to measure the coverage of this relation in Medline.
From the Wnt pathway shown here, we selected four interaction pairs (denoted A–D) and used both concepts of the pair together with Wnt to query EBIMed. The sentences containing both terms (second number, e.g. 3 in A:2/3) from the retrieved Medline abstracts (first number) were used to assess whether the relation between the two proteins is a true interaction as indicated in the diagram.
4.2 Assessment procedure: results
All four analyses provide data that allows for the assessment of the precision of the term identification. Analysis-2, -3 and -4 were used to measure the precision of the identification of meaningful relations. For selected protein–protein relations from the Wnt pathway we assessed the amount of information retrieved by EBIMed in analysis-4.
4.2.1 Term identification is covered by all four analyses (Table 1)
Term identification is a complex task and therefore the results vary for the identification of different types of terms (Hirschman et al., 2005). In all analyses the correct UniProtKB/Swiss-Prot protein was identified at a precision of >90%. Nested protein names were counted as correct, whenever the complete term still referred to the contained protein name. This was the case, if it was followed by a qualifier such as ‘gene’, ‘ortholog’, ‘promotor’, ‘pathway’ or ‘signal’ (e.g. ‘HZF-1’ in ‘HZF-1 orthologue’ was accepted whereas ER in ‘er-ko mice‘ was rejected). The highest identification rate was achieved in analysis-4 (100%) and changed little, when nested terms were excluded. The high precision can be explained by the fact that the selected proteins from the Wnt pathway are not ambiguous in the literature.
We assessed the performance of EBIMed to estimate the precision of term identification in four different analyses
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Type of term . | Term used in correct sense . | Term used in correct sense . | Term incomplete . | Term has other sense . | Total (100%) . |
|---|---|---|---|---|---|---|
| . | . | Nested terms admitted . | Nested terms excluded . | . | . | . |
| Analysis-1: Wnt query. Analysis of first 31 abstracts (31/191) | Protein | 280 (92%) | 165 (54%) | 3 (1%) | 23 (7%) | 306 |
| Species | 91 (76%) | 89 (72%) | 0 | 29 (24%) | 120 | |
| GO term | 83 (95%) | 47 (54%) | 4 (5%) | 0 | 87 | |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (37/94) | Protein | 250 (94%) | 214 (81%) | 12 (5%) | 4 (2%) | 266 |
| Analysis-3: Wnt query. Evaluation of drug–protein relations (37/118) | Protein | 114 (90%) | 99 (78%) | 5 (4%) | 8 (6%) | 127 |
| Drug | 99 (74%) | 62 (47%) | 6 (5%) | 28 (21%) | 133 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (50/110) | Protein | 242 (100%) | 231 (95%) | 0 | 0 | 242 |
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Type of term . | Term used in correct sense . | Term used in correct sense . | Term incomplete . | Term has other sense . | Total (100%) . |
|---|---|---|---|---|---|---|
| . | . | Nested terms admitted . | Nested terms excluded . | . | . | . |
| Analysis-1: Wnt query. Analysis of first 31 abstracts (31/191) | Protein | 280 (92%) | 165 (54%) | 3 (1%) | 23 (7%) | 306 |
| Species | 91 (76%) | 89 (72%) | 0 | 29 (24%) | 120 | |
| GO term | 83 (95%) | 47 (54%) | 4 (5%) | 0 | 87 | |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (37/94) | Protein | 250 (94%) | 214 (81%) | 12 (5%) | 4 (2%) | 266 |
| Analysis-3: Wnt query. Evaluation of drug–protein relations (37/118) | Protein | 114 (90%) | 99 (78%) | 5 (4%) | 8 (6%) | 127 |
| Drug | 99 (74%) | 62 (47%) | 6 (5%) | 28 (21%) | 133 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (50/110) | Protein | 242 (100%) | 231 (95%) | 0 | 0 | 242 |
In analysis-1 (31 abstracts, 191 sentences) the precision for UniProtKB/Swiss-Prot proteins was 92% (recall 94%, not shown). If we did not count nested terms as correct (e.g. HZF-1 nested in ‘HZF-1 orthologue’, grey column), the precision was 54% (recall 55%, not shown). Overall precision for the identification of protein names varied between 90 and 100%. The correct species has been identified at 76% precision. GO terms were identified at 95% precision, mainly due to the fact that exact term matching was applied (Hirschman et al., 2005). (Note: Results in analysis-2 sum up to 101% due to rounding.)
We assessed the performance of EBIMed to estimate the precision of term identification in four different analyses
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Type of term . | Term used in correct sense . | Term used in correct sense . | Term incomplete . | Term has other sense . | Total (100%) . |
|---|---|---|---|---|---|---|
| . | . | Nested terms admitted . | Nested terms excluded . | . | . | . |
| Analysis-1: Wnt query. Analysis of first 31 abstracts (31/191) | Protein | 280 (92%) | 165 (54%) | 3 (1%) | 23 (7%) | 306 |
| Species | 91 (76%) | 89 (72%) | 0 | 29 (24%) | 120 | |
| GO term | 83 (95%) | 47 (54%) | 4 (5%) | 0 | 87 | |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (37/94) | Protein | 250 (94%) | 214 (81%) | 12 (5%) | 4 (2%) | 266 |
| Analysis-3: Wnt query. Evaluation of drug–protein relations (37/118) | Protein | 114 (90%) | 99 (78%) | 5 (4%) | 8 (6%) | 127 |
| Drug | 99 (74%) | 62 (47%) | 6 (5%) | 28 (21%) | 133 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (50/110) | Protein | 242 (100%) | 231 (95%) | 0 | 0 | 242 |
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Type of term . | Term used in correct sense . | Term used in correct sense . | Term incomplete . | Term has other sense . | Total (100%) . |
|---|---|---|---|---|---|---|
| . | . | Nested terms admitted . | Nested terms excluded . | . | . | . |
| Analysis-1: Wnt query. Analysis of first 31 abstracts (31/191) | Protein | 280 (92%) | 165 (54%) | 3 (1%) | 23 (7%) | 306 |
| Species | 91 (76%) | 89 (72%) | 0 | 29 (24%) | 120 | |
| GO term | 83 (95%) | 47 (54%) | 4 (5%) | 0 | 87 | |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (37/94) | Protein | 250 (94%) | 214 (81%) | 12 (5%) | 4 (2%) | 266 |
| Analysis-3: Wnt query. Evaluation of drug–protein relations (37/118) | Protein | 114 (90%) | 99 (78%) | 5 (4%) | 8 (6%) | 127 |
| Drug | 99 (74%) | 62 (47%) | 6 (5%) | 28 (21%) | 133 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (50/110) | Protein | 242 (100%) | 231 (95%) | 0 | 0 | 242 |
In analysis-1 (31 abstracts, 191 sentences) the precision for UniProtKB/Swiss-Prot proteins was 92% (recall 94%, not shown). If we did not count nested terms as correct (e.g. HZF-1 nested in ‘HZF-1 orthologue’, grey column), the precision was 54% (recall 55%, not shown). Overall precision for the identification of protein names varied between 90 and 100%. The correct species has been identified at 76% precision. GO terms were identified at 95% precision, mainly due to the fact that exact term matching was applied (Hirschman et al., 2005). (Note: Results in analysis-2 sum up to 101% due to rounding.)
4.2.2 Relation identification is covered by analysis-2–4
EBIMed is designed to identify proteins that are involved in the same biological process. An example of a biological process is the interaction of two proteins. We therefore estimated the frequency of finding an interaction in a sentence that contains co-occurring proteins. In analysis-2, 40% of the sentences were reporting on a protein–protein interaction (Table 2) and 37% in analysis-4. The numbers were 25 and 34%, respectively, when nested terms were not counted as correct. Failure in the relation identification in analysis-2 and -4 mainly resulted from the use of both terms in parallel sentence structures: either the two proteins appear in a coordination [e.g. ‘… and decreased LRP5 and Dkk-1’ (PMID 15962290)] or in another type of parallel syntactical structure [e.g. ‘Dkk1, but not the related Dkk3, binds LRP6 …’ (PMID 15694380)].
In the analysis-2 and -4 we estimated the rate of a meaningful relations between proteins in sentences with co-occurring UniProtKB/Swiss-Prot protein names (40% in analysis-2, row 1; 37% in analysis-4, row 3)
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Interaction shown . | Interaction shown . | Other type of relation . | Both terms have similar function . | Similar effects by drug vs. protein . | Effect of drug protein . | Parallel syntax . | No relation . | Total (100%) . |
|---|---|---|---|---|---|---|---|---|---|
| . | Nested terms admitted . | Nested terms excluded . | . | . | . | . | . | . | . |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (24/33) | 52 (40%) | 32 (25%) | 10 (8%) | 4 (3%) | 46 (36%) | 17 (13%) | 129 | ||
| Analysis-3: Wnt query. Evaluation of drug–protein relations (19/31) | 49 (50%) | 25 (25%) | 14 (14%) | 3 (3%) | 11 (11%) | 3 (3%) | 18 (18%) | 98 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (26/43) | 46 (37%) | 43 (34 %) | 5 (4%) | 5 (4%) | 61 (48%) | 9 (7%) | 126 |
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Interaction shown . | Interaction shown . | Other type of relation . | Both terms have similar function . | Similar effects by drug vs. protein . | Effect of drug protein . | Parallel syntax . | No relation . | Total (100%) . |
|---|---|---|---|---|---|---|---|---|---|
| . | Nested terms admitted . | Nested terms excluded . | . | . | . | . | . | . | . |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (24/33) | 52 (40%) | 32 (25%) | 10 (8%) | 4 (3%) | 46 (36%) | 17 (13%) | 129 | ||
| Analysis-3: Wnt query. Evaluation of drug–protein relations (19/31) | 49 (50%) | 25 (25%) | 14 (14%) | 3 (3%) | 11 (11%) | 3 (3%) | 18 (18%) | 98 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (26/43) | 46 (37%) | 43 (34 %) | 5 (4%) | 5 (4%) | 61 (48%) | 9 (7%) | 126 |
The same analysis for drug–protein relations (e.g. activation, inhibition, upregulation or downregulation of the protein; analysis-3) lead to the precision of 50% (row 2). Figures were lower, if nested terms were not counted as correct (grey column). Columns 3–6 describe the frequency of several types of co-occurrences encountered that do not describe protein–protein or drug–protein relations. (Note: Results in analysis-3 sum up to 99% due to rounding.)
In the analysis-2 and -4 we estimated the rate of a meaningful relations between proteins in sentences with co-occurring UniProtKB/Swiss-Prot protein names (40% in analysis-2, row 1; 37% in analysis-4, row 3)
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Interaction shown . | Interaction shown . | Other type of relation . | Both terms have similar function . | Similar effects by drug vs. protein . | Effect of drug protein . | Parallel syntax . | No relation . | Total (100%) . |
|---|---|---|---|---|---|---|---|---|---|
| . | Nested terms admitted . | Nested terms excluded . | . | . | . | . | . | . | . |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (24/33) | 52 (40%) | 32 (25%) | 10 (8%) | 4 (3%) | 46 (36%) | 17 (13%) | 129 | ||
| Analysis-3: Wnt query. Evaluation of drug–protein relations (19/31) | 49 (50%) | 25 (25%) | 14 (14%) | 3 (3%) | 11 (11%) | 3 (3%) | 18 (18%) | 98 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (26/43) | 46 (37%) | 43 (34 %) | 5 (4%) | 5 (4%) | 61 (48%) | 9 (7%) | 126 |
| Type of analysis (no. of abstracts containing sentences/no. of unique sentences analysed) . | Interaction shown . | Interaction shown . | Other type of relation . | Both terms have similar function . | Similar effects by drug vs. protein . | Effect of drug protein . | Parallel syntax . | No relation . | Total (100%) . |
|---|---|---|---|---|---|---|---|---|---|
| . | Nested terms admitted . | Nested terms excluded . | . | . | . | . | . | . | . |
| Analysis-2: Wnt query. Evaluation of protein–protein relations (24/33) | 52 (40%) | 32 (25%) | 10 (8%) | 4 (3%) | 46 (36%) | 17 (13%) | 129 | ||
| Analysis-3: Wnt query. Evaluation of drug–protein relations (19/31) | 49 (50%) | 25 (25%) | 14 (14%) | 3 (3%) | 11 (11%) | 3 (3%) | 18 (18%) | 98 | |
| Analysis-4: Evaluation of protein–protein relations from the Wnt pathway (26/43) | 46 (37%) | 43 (34 %) | 5 (4%) | 5 (4%) | 61 (48%) | 9 (7%) | 126 |
The same analysis for drug–protein relations (e.g. activation, inhibition, upregulation or downregulation of the protein; analysis-3) lead to the precision of 50% (row 2). Figures were lower, if nested terms were not counted as correct (grey column). Columns 3–6 describe the frequency of several types of co-occurrences encountered that do not describe protein–protein or drug–protein relations. (Note: Results in analysis-3 sum up to 99% due to rounding.)
Fifty per cent of the co-occurrence of a drug name with a UniProtKB/Swiss-Prot protein name in analysis-3 led to a finding where the drug explicitly had an effect on the protein, e.g. inhibition of the protein or upregulation or downregulation; 25% if nested terms were not counted as correct. Altogether at least one in four sentences containing a protein–protein pair or a protein–drug pair reports on a meaningful relation.
4.2.3 Coverage of relation identification in analysis-4
Querying EBIMed with Wnt retrieved sentences for 74 protein–protein pairs out of the total of 108 pairs described either in the KEGG or the STKE pathway. This included all 10 pairs common to both pathways (results not shown). Failure to retrieve pairs was due to several reasons. First, 13 concepts from the pathway resources are not supported by names and synonyms from the UniProtKB/Swiss-Prot resource, e.g. Diversin, which lead to 21 pairs that could not be verified by EBIMed. Second, for 11 pairs the concepts used were supported by UniProtKB/Swiss-Prot, but the abstracts did not contain sentences with any pair referring to them. Last, in three cases KEGG and STKE use a generalization instead of the more specific terms from UniProtKB/Swiss-Prot (Dkk versus Dkk-1), which were then missed.
For all selected interaction pairs (A–D) in Figure 2, EBIMed retrieved statements that confirmed the selected interaction. For APC and PP2A, three sentences were retrieved and all confirm the interaction between both proteins (interaction, dephosphorylation and complex formation) (PMID 10862053: Webster et al., 2000; PMID 10092233: Seeling et al., 1999). For NLK and TAK1, five sentences report an activation of NLK by TAK1 (PMID 12482967: Ishitani Tohru et al., 2003; PMID 15082531: Kanei-Ishii Chie et al., 2004; PMID 12047350: Hyodo-Miura Junko et al., 2002; PMID 10683140: Behrens, 2000; and PMID 10391247: Shitani et al., 1999) and in 7 cases the authors mention the TAK1-NLK Pathway (remaining 13 cases: both act on a third agent). For GSK-3 versus beta-catenin, EBIMed returned 233 abstracts of which 24 were curated containing 38 statements. Seven cases reported a relationship between the two interaction partners: ‘phosphorylation’ (5), ‘inhibits’ (1) and ‘complex formation’ (1).
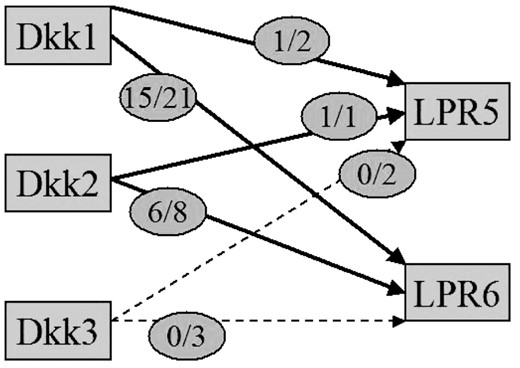
In the case of the interaction pair Dkk and LRP, the retrieved sentences gave a more detailed picture than the one known from the public pathway representations (Fig. 3). 21 sentences (out of 34) indicate that Dkk-1 binds to LRP6, out of which six indicate its inhibition. Four sentences suggest that Dkk-2 interacts with LRP6, two findings suggesting activation. In the case of LRP5, five findings for a relation suggest that Dkk-1 binds to LRP5 and inhibits it. One sentence states that Dkk-2 is a ligand to LRP5. No confirmation was found that Dkk-3 interacts with LRP5 or LRP6.
Querying EBIMed for Dkk AND LRP leads to the retrieval of abstracts with statements describing the relationship between subtypes of LRP and Dkk (43 sentences, 16 abstracts), which gives a more detailed picture than the KEGG representation (Fig. 2). The numbers in the circles represent the total number of sentences with the protein pair in (2nd number) and the number of sentences with a meaningful interaction (1st number). Dotted lines indicate that the retrieved sentences do not contain any indication of a relation between Dkk-3 and LRP5 and LRP6.
To summarize EBIMed extracts protein names at a rate of >90% precision, while 37% of the extracted protein pairs and 50% of the drug–protein pairs represent a meaningful interaction. For the interaction pair Dkk and LRP of the Wnt pathway we were able to clarify the interactions between the subtypes of Dkk and LRP, which are neither documented in KEGG nor in STKE.
Altogether, the use of EBIMed leads to a better access to statements in Medline in comparison to PubMed because the user reads mainly relevant sentences. In the case of the four evaluation examples, 110 sentences were automatically selected from 52 abstracts. 43 out of the 110 sentences carried relevant information (39% precision), which did not require the user to read the remaining information in the abstract. This is an improvement of 16% over the baseline precision of 13% that results from completely reading all 52 abstracts.4
5 DISCUSSION AND CONCLUSION
EBIMed filters Medline abstracts for information such as biomedical terminology and assembles a table containing all encountered pairs. Every entry of the table links to the list of sentences belonging to this pair; every sentence is linked to its abstract; and every identified term in a sentence or an abstract is linked to the terminological resource from which it was extracted. Prioritization of pairs according to the quantity of evidence supporting the pair helps the user to focus on the most relevant concepts and thus reduces reading. Altogether EBIMed organizes the extracted information into a hypermatrix of terms, pairs and sentences leading to the retrieved abstracts.
The advantages of EBIMed are based on three important design principles as follows: (1) the keyword query is independent of the analysis provided by EBIMed, (2) EBIMed generates links between resources and (3) EBIMed extracts, ranks and sorts single sentences as the key information source. In contrast to iHOP and BioIE, EBIMed makes extensive use of biomedical terminological resources and processes several thousand Medline abstracts upon request (Hoffmann and Valencia, 2004; Divoli and Attwood, 2005). Other solutions have been proposed that integrate ontologies for text mining, e.g. Textpresso and GoPubMed. In the first case the set of ontologies comprises 33 categories of concepts that have been developed to curate the literature on Caenorhabditis elegans for the WormBase project (Author Webpage) and therefore forms a solution suitable to members of the WormBase project team (Muller et al., 2004). GoPubMed incorporates GO only and allows to browse Medline abstracts with the help of GO annotations to single abstracts, but does not incorporate any other terminologies (Doms and Schroeder, 2005). In EBIMed we link terminological findings such as GO terms, drugs and species to UniProtKB/Swiss-Prot protein names. This processing step is basic to GO annotation of proteins, the identification of drug targets and species identification for a protein, respectively.
The advantages of curated controlled vocabularies and ontologies is that they have the consensus of a larger user community (e.g. GO), are attached to relevant domain knowledge (e.g. UniProtKB/Swiss-Prot protein names to data entries in UniProt) and develop into comprehensive terminological resources over time. However, they reduce the recall if they are not yet complete or differ from the use of terminology in the scientific literature (e.g. morphological or syntactical term variability in the text). Fundel et al. defined rules for synonymy relations in the case of varying protein names and thus extended existing synonym lists for mouse and yeast protein names to generate higher recall (e.g. considering an extension ‘p’ in SOH6p as synonymous to SOH6) (Fundel et al., 2005). Such rules might be misleading if used for names of other species. Therefore, EBIMed is designed to be fully compliant to public resources and will contribute to the harmonization of both resources in the future. Examples for improvement are protein names such as ‘embryonic’ (UniProt accession No. P02301) and ‘Proteins 5’ (P10463). Regarding the other terminologies, disambiguation of species (‘cancer’, ‘axis’, ‘beta’, ‘idea’) and consistent resources on drugs will contribute to better information extraction and retrieval.
EBIMed leads the user to sentences where proteins have been reported in the same biological process. A small subset is the co-occurrences of protein names that report an interaction (25–50% of the sentences). We conclude that EBIMed's selection of sentences allows faster identification of facts from Medline abstracts than reading one abstract after the other. Furthermore, EBIMed supports other use scenarios not described above such as the identification of drug-related targets. For example, the query Viagra leads to the retrieval of the pair Viagra and phosphodiesterase (PDE5) with the largest number of findings amongst the UniProtKB/Swiss-Prot terms. And the PDE5 is indeed the drug target of Viagra.
Finally, it should be noted that EBIMed fulfils tasks that are complementary to PubMed's use cases. PubMed offers users to tune their queries to their needs. In a restrictive query only a small number of abstracts is returned at all and in a general query the most recent publications cover the query topic well. These two settings are, however, not among the expected use cases for EBIMed. In contrast, EBIMed's table may not be very helpful for a small number of abstracts and for any information that is redundantly mentioned in a large set of abstracts it may not improve access.
EBIMed's tables support users to get an overview on a multitude of relations spread over many abstracts. While individual needs may differ, it is unlikely that a user will examine many hundred abstracts in order to get an overview, for example, how a selection of drugs have been applied in the context of proteins. Access to individual statements about these drugs and proteins is much easier when starting from the EBIMed table than, for example, by submitting a query per drug. Furthermore, the individual would require for this analysis a complete collection of drug names which might not be available. As a result the individual would profit from EBIMed's analysis, which automatically generates the complete list from the encountered findings. Altogether EBIMed allows to use general queries for the retrieval of relations instead of querying PubMed for specific pairs one at a time.
In the future we want to extend EBIMed in several directions. EBIMed has the potential to exploit more advanced literature analysis modules such as syntactical identification of relations, which is already in use by another module processing pipeline, Whatizit (Author Webpage) (Kirsch et al., 2006). Disambiguation of terms as part of EBIMed is ongoing research work. Integration of full paper and other types of documents such as patent abstracts will lead to a more comprehensive retrieval of information.
EBIMed has the potential to induce a paradigm shift towards a situation where authors express explicitly the facts that they want to convey to their audience. The right choice of terminology and the precise phrasing leads to immediate access to the sentences that contain the key facts of a publication in a format that can be interpreted by computers to save the researcher time and effort.
Network of Excellence ‘Semantic Interoperability and Data Mining in Biomedicine’ (NoE 507505). Medline abstracts are provided from the NLM (Bethesda, MD, USA) and PubMed (Author Webpage) is the premier Web portal to access the data. Sylvain Gaudan is supported by an ‘E-STAR’ fellowship funded by the EC's FP6 Marie Curie Host fellowship for Early Stage Research Training under contract number MESTCT-2004-504640.
REFERENCES
1Note that PubMed is National Library of Medicine's (NLM's) interface to allow access to all Medline abstracts. PubMed provides access to more abstracts than contained in the Medline distribution delivered from the NLM to the EBI (e.g. citations in progress and a few additional journals are not part of the Medline distribution).
2percentage of correct findings amongst all findings by a method (= 100 * true positives/(true positives + false positives).
3percentage of facts correctly identified by a method amongst all facts mentioned in the text (= 100 * true positives/(true positives + false negatives).
4On average a Medline abstract contains 6.16 sentences (191 sentences in 31 abstracts, Table 1), which leads to 320 sentences in 52 Medline abstracts. The user has to read the complete abstract to have 100% recall on all UniProtKB/Swiss-Prot protein co-occurrences and interactions. This leads to the result that the user has to read 320 sentences, to find all 43 identified sentences. This results to 13% precision..