The anaplastic lymphoma kinase (ALK) gene regulates cell growth and plays an essential role in the development of the brain by helping with the proliferation of nerve cells1. ALK becomes oncogenic when it acquires gene-specific mutations, forms a fusion gene with other genes, or gains additional gene copies. This ALK genetic abnormality is a key oncogenic driver, especially in non-small cell lung cancer (NSCLC) where it accounts for 3–7% of NSCLC cases in the United States2,3,4.
Recent studies show that tumor cells derived from a subset of patients with NSCLC harbor the echinoderm microtubule-associated protein-like 4 (EML4) fusion oncogene, which is the result of a paracentric chromosomal inversion on the short arm of chromosome 2. The EML4-ALK oncogene, like other ALK fusion oncogenes, is a druggable target that is responsive to ALK inhibitors. However, there is a lack of EML4-ALK in vitro models for drug screening. Here, we set out to generate an isogenic EML4-ALK fusion NSCLC model in the A549 lung cancer cell line (ATCC® CCL-185IG™), which contains other naturally occurring genomic aberrations inherent in NSCLC. This model could serve as a clinically relevant drug screening cell model5,6.
Results and Discussion
Gene editing with CRISPR/Cas9
We employed the CRISPR/Cas9 genome editing platform for the generation of the desired targeted genomic rearrangement in the A549 lung cancer cell line. Single guide RNAs (sgRNAs) designed and built to guide Cas9 to bind and cut desired intronic regions in the EML4 and ALK gene targets, trigger the paracentric genomic rearrangement event upon co-transfection (Figure 1).
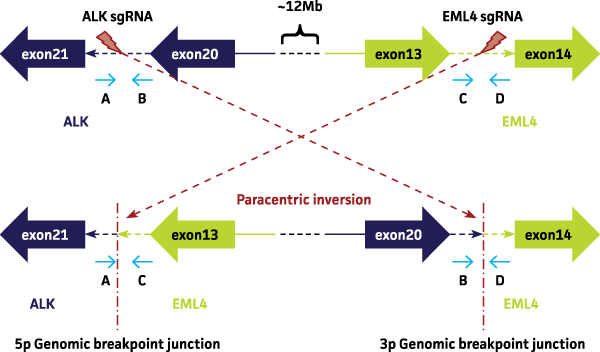
Figure 1. Identification of sgRNA target sites at EML4 and ALK genomic loci. SgRNAs designed and built to guide Cas9 to bind and cut desired intronic regions in the EML4 and ALK gene targets can trigger the paracentric genomic rearrangement event upon cotransfection. ‘A’, ‘B’, ‘C’ and ‘D’ denote primer locations. Transfected cells are sorted into single cells and expanded for screening of gene translocation events by junction PCR across the 5 prime (5p) and 3 prime (3p) genomic breakpoint junctions as shown. The occurrence of a positive ~500bp PCR band for any isolated single cell clone using the primers ‘A’ and ‘C’ as well as ‘B’ and ‘D’ suggests the successful generation of an EML4-ALK fusion product.
Genotype of EML4-ALK mutated cell line
The introduction of the EML4-ALK mutation in the cell line was confirmed via Sanger sequencing as shown in Figure 2 (A, B) for the expected 5p and 3p genomic breakpoints. Sanger sequencing of prepared EML4-ALK cDNA from mRNA of the mutated cell line was carried out to confirm the expression of the EML4-ALK fusion transcript (Figure 2C). We subsequently confirmed expression of the EML4-ALK fusion protein to be an 86 kDa fragment as expected by western blotting (data not shown).
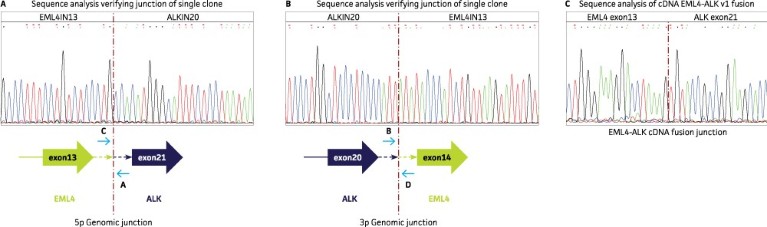
Figure 2. (A, B) Sanger sequencing results for the 5p and 3p genomic junction PCR confirmed the accuracy of the generated EML4-ALK fusion product. Shown below each chromatogram is a cartoon representation of the edited genomic locus. The red dashed line indicates the genomic fusion junction for the ALK and EML4 genes. (C) Sequence of EML4-ALK fusion transcript across cDNA breakpoint for the isolated clone. The cyan dashed line is the EML4-ALK cDNA fusion junction.
Functional characterization of EML4-ALK mutated cell lines
Functional testing of the isogenic A549 EML4-ALK cell line gave a favourable drug response in comparison to its parental A549 cell line (Figure 3). Dose response curves for cells treated with ALK inhibitors crizotinib and ceritinib (Figure 3 A, B) showed that the isogenic A549 EML4-ALK cell line has selective drug sensitivity to ALK inhibitors crizotinib and ceritinib relative to the parental A549 cell line. Furthermore, this trend is consistent regardless of whether it is a dose-response based assay monitored via IncuCyte FLR live cell imaging system (Essen BioScience; Figure 3 A, B) or the CellTiter-Glo® lumniscent cell viability assay (Promega; Figure 3 C, D).
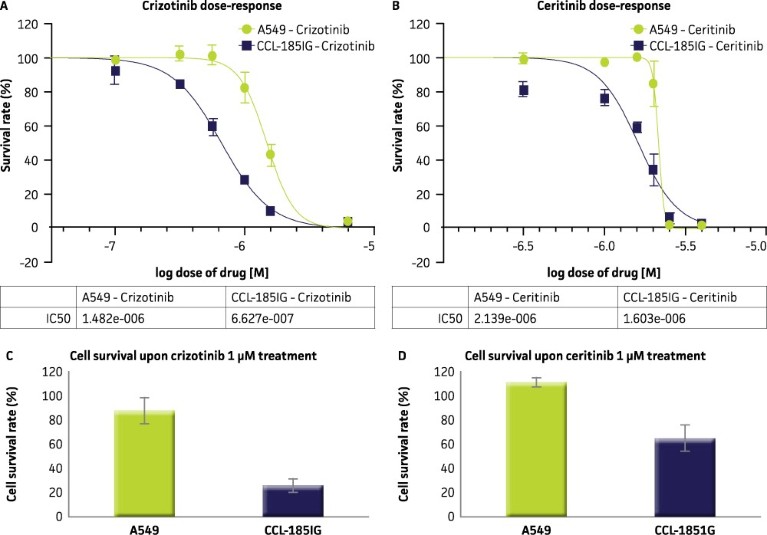
Figure 3. CCL-185IG is sensitive to ALK inhibitor drugs. (A, B) A549 and CCL-185IG cells were treated with the indicated concentrations of ALK inhibitors crizotinib and ceritinib, and cell survival was determined via live cell analysis. (C, D) A549 and CCL-185IG cells were treated with 1 µM of same compounds and cell survival was confirmed by CellTiter-Glo® luminescent cell viability assay.
Conclusion
In this study, we used the CRISPR/Cas9 genome editing platform to target endogenous loci in human cells and create the intended genomic translocation event. By employing sgRNA-Cas9 constructs designed to cut precisely at relevant translocation breakpoints, we induced cancer-relevant genomic rearrangements that resulted in the expression of EML4-ALK gene fusion products. Breakpoint junction analysis tested after sgRNA-CRISPR/Cas9-mediated genomic DNA cleavage in A549 cells revealed the successful creation of the EML4-ALK fusion found in tumor cells from a subpopulation of NSCLC patients. Furthermore, single clonal isolation and functional screening demonstrated that the EML4-ALK isogenic cell line (ATCC® CCL-185IG™) was sensitive to ALK inhibitors relative to the parental A549 cell line. This newly developed EML4-ALK isogenic lung cancer cell line is a useful model to study the tyrosine kinase signaling pathway and to screen for novel ALK inhibitors in anti-cancer drug discovery and development.


 Cutting edge resources to accelerate innovative research
Cutting edge resources to accelerate innovative research