- 1Nanhua Hospital Affiliated to University of South China, Hengyang, China
- 2Transformation Research Lab, Hengyang Medical School, University of South China, Hengyang, China
- 3The Third Affiliated Hospital of University of South China, Hengyang, China
Tamoxifen is a drug commonly used in the treatment of breast cancer, especially for postmenopausal patients. However, its efficacy is limited by the development of drug resistance. Downregulation of estrogen receptor alpha (ERα) is an important mechanism of tamoxifen resistance. In recent years, with progress in research into the protective autophagy of drug-resistant cells and cell cycle regulators, major breakthroughs have been made in research on tamoxifen resistance. For a better understanding of the mechanism of tamoxifen resistance, protective autophagy, cell cycle regulators, and some transcription factors and enzymes regulating the expression of the estrogen receptor are summarized in this review. In addition, recent progress in reducing resistance to tamoxifen is reviewed. Finally, we discuss the possible research directions into tamoxifen resistance in the future to provide assistance for the clinical treatment of breast cancer.
Introduction
Breast cancer is the most common cancer in women (Bray et al., 2018), and endocrine therapy plays an important role in breast cancer treatment (Rugo et al., 2016). More than 60% of breast cancers are estrogen-receptor (ER) positive (Lopez-Tarruella and Schiff, 2007; Vargo-Gogola and Rosen, 2007). Tamoxifen is an antagonist of ERα66, and it is commonly used in the treatment of ER-positive breast cancers (Binkhorst et al., 2012); however, the efficacy is not satisfactory because of the development of tamoxifen resistance. RTKs (receptor tyrosine kinases) and the activation of the PI3K-PTEN/AKT/mTOR pathway caused by the overexpression of RTKs are thought to be closely related to resistance to tamoxifen (Hosford and Miller, 2014; Yin et al., 2014).
On the other hand, ERα36, a 36 kDa truncated isoform of ERα66 located on the cytoplasmic membrane of breast cancer (Lv et al., 2015; Omarjee et al., 2017), has been reported to be related to the drug resistance and metastasis of cancer cells (Zhang and Wang, 2013; Yin et al., 2014; Omarjee et al., 2017). Tamoxifen can activate ERα36, which in turn activates MAPK, AKT, and other signaling pathways, leading to tamoxifen resistance (Tong et al., 2010).
In recent years, a large body of evidence has shown that protective autophagy, cell cycle regulators, and some transcription factors play a key role in tamoxifen resistance, such as KLF4 regulating drug resistance by regulating MAPK and the discovery of LEM4 (Gao et al., 2018; Jia et al., 2018). Scientists have proposed many methods to reduce drug resistance through these mechanisms and have made great progress.
In this review, the development of tamoxifen resistance in breast cancer is discussed, with special emphasis on the effects of some newly discovered enzymes and transcription factors on tamoxifen resistance, the protective autophagy of cells, and the latest progress in cell cycle regulators.
The Role of Receptor Tyrosine Kinases (RTKs) in Tamoxifen Resistance
RPTKs are a class of enzyme-linked receptors that have been found to come in many kinds, including epidermal growth factor (EGF) receptor, platelet-derived growth factor (PDGF) receptor, macrophage colony stimulating factor (M-CSF), insulin and insulin-like growth factor-1 (IGF-1) receptor, vascular endothelial growth factor (VEGF) receptor, and hepatocyte growth factor (HGF) receptor. The PI3K/AKT/mTOR signaling pathway is one of the important mechanisms of tamoxifen resistance, and HER2 activates PI3K as a member of the EGFR family (Mansouri et al., 2018a). It has been proven that high expression of p-AKT is associated with a worse prognosis, and inhibiting the expression of AKT is beneficial for sensitizing drug-resistant cells (Block et al., 2012; Karlsson et al., 2019). In addition, activation of the PI3K/AKT pathway is not just associated with tamoxifen resistance. Recent studies have shown that activation of the PI3K/AKT pathway can cause tamoxifen-resistant (TAM-R) cells to develop drug resistance to DNA-damaging chemotherapy by upregulating BARD1 and BRCA1 (Zhu et al., 2018), which makes the PI3K/AKT pathway particularly important in the treatment of breast cancer.
The mechanism of activation of the PI3K/AKT/mTOR pathway has also been studied by many scientists. CC chemokine ligand 2 (CCL2), which is secreted by tumor-associated macrophages (TAMs), has been found to be related to activation of the PI3K/AKT/mTOR pathway. However, NF-κB promotes the secretion of CCL2 (Li et al., 2020a). Inhibition of the PI3K/AKT pathway may be beneficial to improve the efficacy of chemotherapy and endocrine therapy for breast cancer patients. Many drugs targeting PI3K, mTOR, or AKT to overcome tamoxifen resistance have been put into use. However, due to the complexity of the PI3K/AKT/mTOR pathway, inhibiting the pathway at any level will activate compensatory mechanisms, which limits the efficacy of inhibitors (Choi et al., 2016; Lui et al., 2016). We need to study the cross-talk between these pathways in future research.
The combined use of several inhibitors may be an important way to improve tamoxifen resistance in the future. Both VEGF and HER2 are members of the RTK family. Studies have shown that the expression of VEGF in drug-resistant cells is upregulated. VEGF contributes to angiogenesis and promotes tumor growth, which is not conducive to a good prognosis of breast cancer patients (Oh et al., 2010). The MAPK/ERK pathway has been proven to contribute to tamoxifen resistance (Heckler et al., 2014; Peng et al., 2017; Yin et al., 2017), whereas VEGF overexpression in drug-resistant cells leads to increased activation of MAPK. Surprisingly, the use of VEGF inhibitors was not found to be helpful in overcoming tamoxifen resistance (Mansouri et al., 2018b), which may also be attributed to the complex network of drug resistance. There is still no evidence that VEGF is related to tamoxifen resistance.
EGFR is also thought to be related to tamoxifen resistance. Tamoxifen downregulates the expression of miR-186-3p, which leads to further upregulation of the expression of EREG, a target gene of miR-186-3p. EREG then activates EGFR even more, subsequently enhancing glycolysis and leading to tamoxifen resistance (He et al., 2019). It has been reported that the NOGO-B receptor is related to tamoxifen resistance. The NOGO-B receptor contributes to the transport of RAS, which enhances EGF signal transduction, resulting in a decrease in p53 expression and the development of drug resistance (Gao et al., 2018).
ERα36 has been reported to be associated with tamoxifen resistance (Yin et al., 2014), and ERα36 reduces the sensitivity of breast cancer cells to tamoxifen by upregulating EGFR. EGFR expression and the basal level of ERK phosphorylation are upregulated in TAM-R cells. The EGFR/ERK signaling pathway can be blocked by knocking out ERα36 (Li et al., 2020b). However, lapatinib cannot only inhibit the phosphorylation of EFGR and HER2, but also decreases the expression of ERα36 (Yin et al., 2014). Interestingly, studies have shown that cross-talk between HER2 and ERK is conducive to the development of drug resistance (Ito et al., 2012).
In addition to members of the RTK family, such as HER2, EGFR, and VEGF, some research has shown that IGFR is also associated with tamoxifen resistance. Inhibition of IGF-1R reduces the sensitivity of cells to tamoxifen, which may be due to the inhibition of FoxO1 expression by the reduction of IGF-1R expression (Vaziri-Gohar et al., 2017). However, IGF1R signaling may be beneficial to the development of tamoxifen resistance in some aspects. P21-activated kinase 2 (PAK2) is a tamoxifen resistance inducer, while IGF1R can lead to the development of tamoxifen resistance by promoting the expression of PAK2 (Zhang et al., 2018).
In general, there is a complex network in the mechanisms of action of the RTK family, and ERα36 affects the sensitivity of breast cancer cells to tamoxifen. These signaling cascades are described in Figure 1. The development of inhibitors for corresponding targets based on these mechanisms is the focus of previous research. However, due to the compensatory mechanisms that appear when any specific target is inhibited, the clinical effect of improving drug resistance is not very significant. Therefore, studies on improving drug resistance by other mechanisms have emerged in recent years.
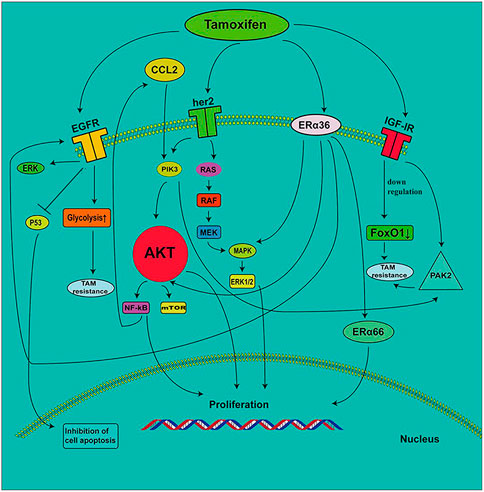
FIGURE 1. The role of RTKs and ERα36 in the development of tamoxifen resistance. EGFR induces tamoxifen resistance by enhancing the glycolytic pathway. The increase in EGF signal transduction induces a decrease in P53 expression, which leads to the inhibition of cell proliferation. TAMs secrete CC-chemokine ligand 2 (CCL2), which activates the PI3K/AKT/mTOR pathway. NF-κB promotes the secretion of CCL2. ERα36 contributes to the upregulation of EGFR, which increases ERK phosphorylation. The decrease in IGF-1R expression leads to the inhibition of FoxO1 expression, which results in the development of tamoxifen resistance. IGF1R mediates the expression of PAK2 and leads to drug resistance.
The Role of Enzymes and Transcription Factors in Tamoxifen Resistance
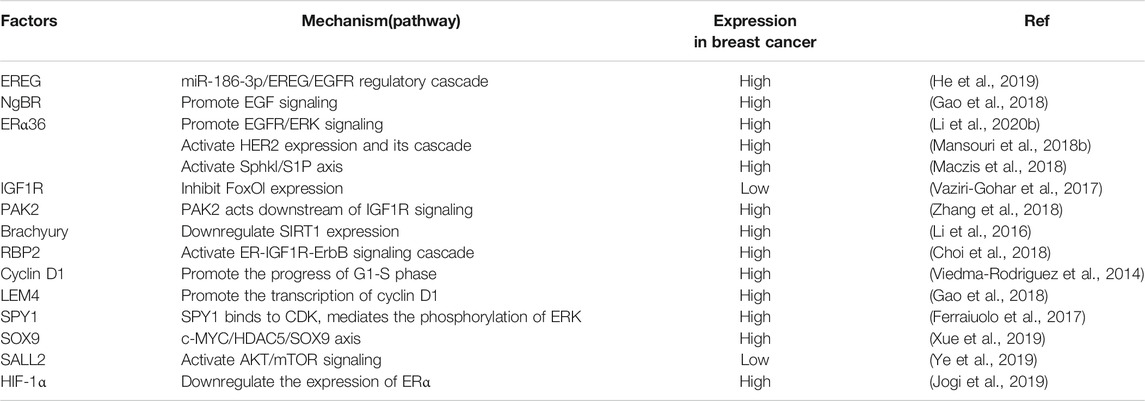
Based on the aforementioned mechanism, some enzymes and transcription factors also play a vital role in the complex network of ER-positive breast cancer resistance to tamoxifen. SOX9 is a transcription factor related to endocrine resistance (Jeselsohn et al., 2017; Xue et al., 2019). Histone deacetylase 5 (HDAC5), a member of the HDAC family whose main function is to remove acetyl groups, enables the deacetylation of SOX9 to facilitate its nuclear localization in TAM-R cells. Moreover, MYC plays an important role in the activation of HDAC5 transcription, and the C-MYC/HDAC5/SOX9 axis is related to tamoxifen resistance (Xue et al., 2019).
HDAC1, another member of the HDAC family, has also been reported to be associated with tamoxifen resistance. The expression of RBP2 is significantly higher in TAM-R cells than in cells sensitive to tamoxifen. The RBP2–ER–NRIP1–HDAC1 complex leads to IGF1R activation. The relationship between RBP and tamoxifen resistance is related to the PI3K/AKT pathway. RBP activates the PI3K/AKT pathway by enhancing the cross-talk between IGF1R and the HER2 receptor, which leads to drug resistance (Choi et al., 2018). Interestingly, it has also been reported that HDAC promotes the expression of ERα66 and AKT, and the use of HDAC inhibitors can inhibit the level of AKT by reducing the stability of its mRNA (Thomas et al., 2013).
Silent information regulator 2-related enzyme 1 (SIRT1) is a deacetylase dependent on nicotinamide adenine dinucleotide, which is highly expressed in a variety of tumors and has been proven to inhibit the growth of breast cancer cells (Liu et al., 2009; Kuo et al., 2013). The T-box protein Brachyury, a transcription factor, promotes the resistance of breast cancer cells to tamoxifen by inhibiting SIRT1 (Li et al., 2016). There are many different mechanisms for the effects of acetylases on tamoxifen resistance.
Estrogen regulates tumor growth by binding to ERα66 in the cytoplasm. Tamoxifen is antagonistic to ERα66. However, the use of tamoxifen has been confirmed to be involved in the upregulation of ERα36, SPhk1 (sphingosine kinase 1), and S1P (sphingosine-1-phosphate), which further activates downstream signaling pathways and causes drug resistance (Maczis et al., 2018), while the inhibition of ERα36 is beneficial to restore the sensitivity of breast cancer cells to tamoxifen.
Protein arginine N-methyltransferase 2 (PRMT2; HRMT1L1) is a member of the arginine methyltransferase family (Scott et al., 1998) that inhibits the resistance of breast cancer cells to tamoxifen by inhibiting the ERα36, PI3K, MAPK, and other signaling pathways (Shen et al., 2018).
It has been shown that the expression of hypoxia inducible factor HIF-1α contributes to the decrease in ERα, which is related to the sensitivity of endocrine therapy. HIF-1α reduces the sensitivity of breast cancer cells to tamoxifen. Interestingly, the expression of HIF-1α is related to the expression of EGFR (Jogi et al., 2019).
In contrast to HIF-1α, Spalt-like transcription factor 2 (SALL2), a transcription factor related to disease progression, enhances the sensitivity of breast cancer cells to tamoxifen, while ERα is downregulated after silencing SALL2 (Ye et al., 2019). This shows that tamoxifen is an effective endocrine therapy drug in ER-positive breast cancer patients. However, the expression of ERα is positively correlated with the sensitivity of tamoxifen therapy in ER-positive breast cancer patients. Numerous transcription factors regulate the sensitivity of breast cancer cells to tamoxifen by regulating ERα through various mechanisms. In addition, ERα also mediates the expression of glutathione S-transferase mu 3 (GSTM3) to resist cytotoxicity caused by drug therapy and to protect the drug-resistant cells.
The expression of GSTM3 was found to be higher in HER2-positive cancer cells (Lin et al., 2018). This indicates that there may be a relationship between GSTM3 and the RTK pathway, and the mechanism by which enzymes and transcription factors regulate tamoxifen resistance is also closely related to the RTK pathway. Li et al. (2018) found that the ER–c-Src–HER2 complex plays a vital role in tamoxifen resistance, while c-Cbl reverses tamoxifen resistance by inhibiting the formation of the ER–c-Src–HER2 complex. It seems that most enzymes are involved in drug resistance through the RTK pathway.
In addition, some enzymes can be used to predict the sensitivity of endocrine therapy in breast cancer. Shimoda et al. (2017) found that the expression of ASPH was upregulated in tamoxifen-resistant cells, and the upregulation depended on the PI3K and MAPK pathways. The cells with high expression of ASPH were more sensitive to tamoxifen than those with low expression of ASPH, and the results were statistically significant.
Aspartate-b-hydroxylase (ASPH) may also predict the sensitivity of breast cancer cells to tamoxifen (Shimoda et al., 2017). Gwak et al. (2017) also found that the expression of the transcription factor OCT 4 may be related to the poor efficacy of tamoxifen, and its expression level can be used to predict the sensitivity of breast cancer cells to tamoxifen. The mechanisms mentioned in this review related to tamoxifen resistance are summarized in Table 1.
The Discovery of LEM4 and the Association Between Cell Cycle Regulators and Resistance to Tamoxifen
As a competitive antagonist of estradiol, tamoxifen can bind to estrogen receptors in competition with estradiol and form a stable complex, which inhibits the transcription activity of the estrogen receptor and blocks breast cancer cells in G1 phase to inhibit tumor proliferation. However, tamoxifen has little effect on the cell cycle when cells are treated with tamoxifen alone (Cheng et al., 2017). Previous studies have shown that cyclin D1 and cyclin E are essential for the emergence of tamoxifen resistance in breast cancer cells. Cyclin D1 promotes the progression of the G1–S phase, and tamoxifen can reduce the expression of cyclin D1, which is highly expressed in drug-resistant cells (Viedma-Rodriguez et al., 2014). Based on these mechanisms, scientists have previously proposed many methods to overcome drug resistance, such as the cyclin-dependent kinase (CDK) 4/6 inhibitors palbociclib and ribociclib (Finn et al., 2015; Cristofanilli et al., 2016; Hortobagyi et al., 2016).
The latest research in the last 2 years found that LEM4 (LEM structural protein), which is highly expressed in breast cancer-resistant cells, promotes the transcription of cyclin D1 through ligand-independent activation of receptors. Furthermore, LEM4 interacts with CDK 4/6 and Rb to accelerate the G1–S transition (Gao et al., 2018). Therefore, LEM4 reduces the inhibitory effect of tamoxifen on the G1–S phase transition of breast cancer cells. On the other hand, the existence of LEM4 allows the estrogen receptor to undergo ligand-independent activation in the presence of tamoxifen. LEM4 is expected to be a biological index to predict tamoxifen resistance in ER-positive breast cancer, and targeting LEM4 may be a feasible research direction to overcome tamoxifen resistance in the future.
In addition, Yu et al. (2019) found that cell division cycle associated 8 (CDCA8) may be related to tamoxifen resistance. It is highly expressed in drug-resistant cells. After the CACA8 gene was knocked out, the number of drug-resistant cells in the G1 phase increased, and the drug resistance of the cells to tamoxifen decreased (Yu et al., 2019).
Ferraiuolo et al. (2017) discovered another cell cycle protein, Spy1, which mediates the phosphorylation of ERK under the condition of binding to CDK; the increase in its level is related to tamoxifen resistance.
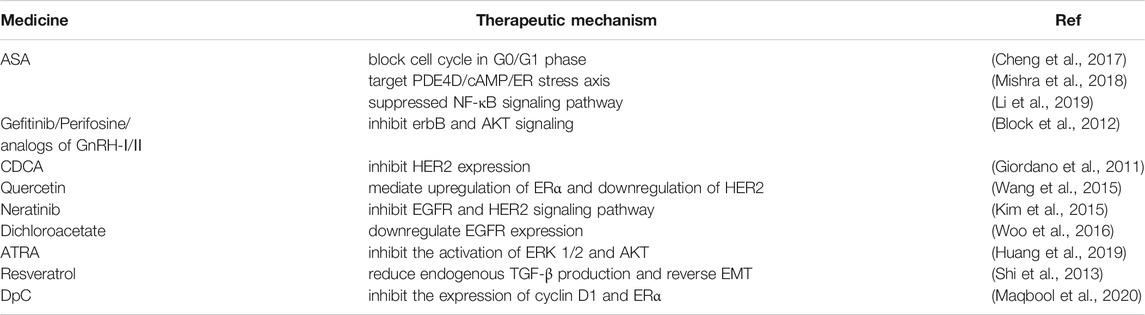
With the progress of mechanistic research, many new treatments have emerged in recent years. Aspirin (ASA) is a kind of nonsteroidal anti-inflammatory drug that has been used in the treatment of many tumors, including rectal cancer, lung cancer, pancreatic cancer, and breast cancer (Jiang et al., 2020; Wang and Huang, 2020; Wu et al., 2020; Zhang et al., 2020), but whether it is beneficial to the survival of patients is still uncertain. However, the use of aspirin seems to be helpful in overcoming tamoxifen resistance. The expression of cyclin D1 was downregulated, and the number of cells arrested in the G0/G1 phase was increased when tamoxifen was used in combination with ASA. The combination of ASA and tamoxifen can overcome the drug resistance of ER-positive breast cancer cells to tamoxifen (Cheng et al., 2017).
Maqbool et al. (2020) synthesized a novel thiosemicarbazone, DpC. They found that the combination of DpC and tamoxifen effectively reduced cyclin D1, upregulated p27, and inhibited the proliferation of breast cancer cells, which may be helpful to overcome the drug resistance of tamoxifen.
The Latest Progress in the Relationship Between Autophagy and Resistance to Tamoxifen
Autophagy is the process by which cells engulf their excess proteins or organelles, transport them to lysosomes, and degrade their contents. Their main role is to deal with the stress-induced injury of cells (Antonioli et al., 2017). However, autophagy seems to have two opposing roles in tumor cells. On the one hand, tumor cells can undergo autophagic cell death through self-phagocytosis, after which the cytoskeleton is mostly preserved. On the other hand, autophagy can delay the apoptosis of stressed and damaged cells, and protect their survival (Cook et al., 2011; Sun et al., 2015). Previous studies have shown that autophagy may have a strong relationship with tamoxifen resistance, and it may be an important mechanism of tamoxifen resistance (Gonzalez-Malerva et al., 2011; Nagelkerke et al., 2014), but the relationship between autophagy and tamoxifen resistance is still in the exploratory stage, and the specific mechanism is still unclear.
Recent studies have suggested that autophagy plays a very important role in cell protection. Lysosome-associated membrane protein (LAMP) is an important mediator of the process of autophagy and lysosome fusion. Autophagy was inhibited, and the cells were re-sensitized to tamoxifen after LAMP3 knockdown (Nagelkerke et al., 2014). TAM-R cells have a higher level of autophagy than tamoxifen-sensitive cells, and inhibition of autophagy will improve the efficacy of TAM (Liu et al., 2019). Wang et al. (2019) found that the expression of the H19 gene was enhanced in TAM-R cells and that H19 was significantly related to the enhancement of autophagy in breast cancer cells. Knockout of the H19 gene could make breast cancer cells re-sensitized to tamoxifen.
Why does tamoxifen enhance autophagy and lead to drug resistance? It is well known that tumor cells need a lot of energy to maintain their growth and proliferation, and a significant amount of this energy comes from enhanced glycolysis (Kim and Dang, 2006). The use of tamoxifen has been found to be related to the energy metabolism of cells. It was found that the ATP level of breast cancer cells decreased after tamoxifen treatment. Moreover, the use of tamoxifen could lead to the upregulation of the expression of MTA1, which further destroys mitochondrial function, while drug-resistant cells meet their energy needs through enhanced autophagy (Lee et al., 2018; Das et al., 2019). We speculate that the enhancement of autophagy may be the result of the increased energy demand of tumor cells and the anti-stress response of tumor cells.
Many autophagy-related genes have been discovered, and many autophagy inhibitors have been developed to inhibit tamoxifen resistance. Cheng et al. (2019) found that the use of icariin significantly increased the apoptosis of TAM-R cells; more TAM-R cells remained in the G0/G1 phase, while S phase/G2 phase cells were significantly reduced. At the same time, the expression of cyclin D1, Bcl-2, LC3-1, LC3-II, AGT5, and Beclin-1 were all downregulated. Interestingly, the expression of Beclin-1 downregulates the estrogen signal, which is beneficial to overcoming the resistance to tamoxifen (John et al., 2008). Similarly, Qi et al. (2017) found that autophagy is beneficial to the survival of breast cancer cells, while Z-ligustilide, which inhibits autophagy, may be helpful to overcome the resistance to tamoxifen in breast cancer.
SEL is an antagonist of XOP1. Combined treatment with SEL and 4-OH tamoxifen downregulated the expression of AKT and activated autophagy by blocking the glycolysis pathway, leading to cell death (Kulkoyluoglu-Cotul et al., 2019). Moreover, the degree of autophagy and the expression of autophagy-related genes can be used to judge drug resistance and select the treatment method, which may be helpful for the treatment of ER-positive breast cancer patients in the future.
The relationship between tamoxifen and energy metabolism may become a key research direction in the future, and it is of great significance to control the apoptosis and proliferation of tumor cells and to restore the sensitivity to tamoxifen.
Progress and Future Direction of Tamoxifen Resistance in Breast Cancer
Endocrine therapy is extremely important for ER-positive breast cancer patients. It mainly includes selective estrogen receptor modulators (SERMs), estrogen receptor downregulated modulators (SERDs), and aromatase inhibitors (AIS). Tamoxifen is one of the SERMs (Ali et al., 2016).
To overcome the resistance to tamoxifen, an increasing number of methods have been studied. ASA can not only reduce drug resistance by blocking G0/G1 phase-resistant cells but also by inhibiting the phosphorylation of AKT (Cheng et al., 2017). Phosphodiesterase 4D (PDE4D) can block cAMP and downstream signaling channels, making cells resistant to tamoxifen. However, the level of cAMP in cells is increased and the phosphorylation level of AKT is decreased after the use of aspirin (Mishra et al., 2018).
In addition, NF-κB has been proven to be related to the resistance of tamoxifen. Li et al. (2019) found that aspirin inhibited the activation of NF-κB signaling, which contributed to overcoming the resistance of cells to targeted therapeutic drugs. Aspirin seems to be a feasible strategy to overcome tamoxifen resistance, and it is expected to provide a new direction to breast cancer treatment. In addition to ASA combined with tamoxifen, proteasome inhibitors (PIs) combined with endocrine therapy have also been proven to be beneficial to the sensitization of tamoxifen-resistant cells (Maynadier et al., 2016; Cheng et al., 2017).
Inhibiting kinases in the RTK pathway to overcome drug resistance is also considered to be a viable approach. For example, gefitinib, perifosine, or GnRH-I and GnRH-II analogs were used to inhibit AKT expression (Block et al., 2012). Giordano et al. (2011) found that the primary bile acid chenodeoxycholic acid (CDCA) can activate the farnesoid X receptor (FXR) and inhibit the expression of HER2. Quercetin has also been found to restore the sensitivity to tamoxifen by mediating the upregulation of ERα and the downregulation of HER-2 (Wang et al., 2015).
The combination of tamoxifen and gefitinib promoted the apoptosis of drug-resistant cells. Gefitinib inhibited the downregulation of ERα by EGFR and restored the sensitivity of cells to tamoxifen to a certain extent (Jeong et al., 2019). Interestingly, another study showed that gefitinib has no effect on the activity of breast cancer-resistant cells, while neratinib, another EGFR inhibitor, induced the apoptosis of resistant cells by inhibiting the EGFR and HER2 signaling pathways (Kim et al., 2015). In addition, the use of dichloroacetate can overcome tamoxifen resistance by downregulating EGFR (Woo et al., 2016). Therefore, further study is needed on the effect of EGFR inhibitors on tamoxifen-resistant cells.
Peptidyl-prolyl isomerase Pin1 participates in the development of drug resistance by inducing E2F-4. Interestingly, all-trans retinoic acid (ATRA), an inhibitor of Pin1, inhibits the drug resistance of cells mainly by inhibiting the ERK 1/2 and AKT pathways (Huang et al., 2019).
Inhibition of epithelial–mesenchymal transition (EMT)-like phenomena is also a direction to take to overcome drug resistance. In addition to LDHA inhibiting EMT-like phenomena, resveratrol can also inhibit EMT by inhibiting TGF-β and overcoming tamoxifen resistance. Interestingly, EGFR activation is also related to EMT-like phenotype change, which confirms that inhibition of EMT contributes to overcome tamoxifen resistance (Zuo et al., 2011; Shi et al., 2013; Das et al., 2019). The drugs that overcome tamoxifen resistance mentioned in this review are summarized in Table 2.
With the advances in science and technology, some new approaches have been developed to improve tamoxifen sensitivity. For example, the application of nanotechnology (Guney Eskiler et al., 2018; Kumar et al., 2018) and the benefits of cold atmospheric plasma (CAP) in overcoming drug resistance, etc. (Lee et al., 2017).
Early judgments about the possible efficacy of endocrine therapy is of great significance in clinical treatment. Therefore, some prognostic markers suggesting tamoxifen resistance have been identified (Putluri et al., 2014; Elias et al., 2015; Gwak et al., 2017; Shimoda et al., 2017; Gong et al., 2018). The discovery of these markers is conducive to making early judgments about endocrine therapy efficacy and predictions of recurrence, which is helpful for doctors when making appropriate changes to the treatment strategy.
Conclusions and Prospects
Tamoxifen plays an important role in ER-positive breast cancer patients. However, drug resistance limits its efficacy, illustrating the importance of overcoming tamoxifen resistance in breast cancer. Most methods to overcome breast cancer resistance are based on the mechanism of drug resistance, such as inhibition of the RTK pathway, upregulation of ERα36, and blocking protective autophagy, cell cycle regulators and EMT-like phenomenon. In addition, some new methods have broadened the field of vision to overcome the drug resistance of tamoxifen. For example, some drugs combined with tamoxifen can inhibit the development of drug resistance, and the development of some new technologies is conducive to reducing the drug resistance of tamoxifen, and some prognostic markers of tamoxifen resistance have been discovered.
Research on the relationship between autophagy, cell cycle regulators, and resistance to tamoxifen has made great progress in recent years. The enhanced autophagy in drug-resistant cells is mainly due to the destruction of mitochondrial function caused by tamoxifen, and drug-resistant cells meet their energy demand through autophagy. The methods to overcome drug resistance according to the autophagy mechanism are mainly limited in the current research to the inhibition of autophagy by autophagy inhibitors.
In addition to continuing to look for better autophagy inhibitors to overcome the resistance, we hypothesized that tamoxifen combined with other drugs that protect mitochondrial function can prevent enhanced autophagy and overcome the drug resistance of tamoxifen. This is a new idea to improve the drug resistance of tamoxifen, and there is little research in this area.
Moreover, by detecting the level of autophagy and the expression of autophagy-related genes, the level of cell resistance can be judged, and treatment can be formulated and changed accordingly, which may improve the clinical treatment of breast cancer.
Targeting LEM4 is a feasible research direction to overcome tamoxifen resistance in the future. It has been proven that the high expression of LEM4 in drug-resistant cells is an important mechanism involved in the attenuation of the inhibitory effect of tamoxifen on the G1–S transition. Targeting LEM4 will play a significant role in overcoming tamoxifen resistance.
Overall, the main direction to overcome tamoxifen resistance in the future is not limited to inhibiting the expression of pathways related to tamoxifen resistance but may focus more on cyclins related to tamoxifen resistance, targeting LEM4 and inhibiting autophagy.
Author Contributions
Conception: JY, JZ, and JH. Collection and assembly of data: All authors; Data analysis and interpretation: JY, JZ, KD, and RZ; Manuscript writing: JY and JZ; Final approval of manuscript: All authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ali, S., Mondal, N., Choudhry, H., Rasool, M., Pushparaj, P. N., Khan, M. A., et al. (2016). Current management strategies in breast cancer by targeting key altered molecular players. Front. Oncol. 6, 45. doi:10.3389/fonc.2016.00045.
Antonioli, M., Di Rienzo, M., Piacentini, M., and Fimia, G. M. (2017). Emerging mechanisms in initiating and terminating autophagy. Trends Biochem. Sci. 42 (1), 28–41. doi:10.1016/j.tibs.2016.09.008.
Binkhorst, L., van Gelder, T., and Mathijssen, R. H. J. (2012). Individualization of tamoxifen treatment for breast carcinoma. Clin. Pharmacol. Ther. 92 (4), 431–433. doi:10.1038/clpt.2012.94.
Block, M., Gründker, C., Fister, S., Kubin, J., Wilkens, L., Mueller, M. D., et al. (2012). Inhibition of the AKT/mTOR and erbB pathways by gefitinib, perifosine and analogs of gonadotropin-releasing hormone I and II to overcome tamoxifen resistance in breast cancer cells. Int. J. Oncol. 41 (5), 1845–1854. doi:10.3892/ijo.2012.1591.
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 68 (6), 394–424. doi:10.3322/caac.21492.
Cheng, R., Liu, Y.-J., Cui, J.-W., Yang, M., Liu, X.-L., Li, P., et al. (2017). Aspirin regulation of c-myc and cyclinD1 proteins to overcome tamoxifen resistance in estrogen receptor-positive breast cancer cells. Oncotarget 8 (18), 30252–30264. doi:10.18632/oncotarget.16325.
Cheng, X., Tan, S., Duan, F., Yuan, Q., Li, Q., and Deng, G. (2019). Icariin induces apoptosis by suppressing autophagy in tamoxifen-resistant breast cancer cell line MCF-7/TAM. Breast Cancer 26 (6), 766–775. doi:10.1007/s12282-019-00980-5.
Choi, A.-R., Kim, J.-H., Woo, Y. H., Cheon, J. H., Kim, H. S., and Yoon, S. (2016). Co-treatment of LY294002 or MK-2206 with AZD5363 attenuates AZD5363-induced increase in the level of phosphorylated AKT. Ar 36 (11), 5849–5858. doi:10.21873/anticanres.11170.
Choi, H.-J., Joo, H.-S., Won, H.-Y., Min, K.-W., Kim, H.-Y., Son, T., et al. (2018). Role of RBP2-induced ER and IGF1R-ErbB signaling in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 110 (4), 400. doi:10.1093/jnci/djx207.
Cook, K. L., Shajahan, A. N., and Clarke, R. (2011). Autophagy and endocrine resistance in breast cancer. Expet Rev. Anticancer Ther. 11 (8), 1283–1294. doi:10.1586/era.11.111.
Cristofanilli, M., Turner, N. C., Bondarenko, I., Ro, J., Im, S.-A., Masuda, N., et al. (2016). Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 17 (4), 425–439. doi:10.1016/S1470-2045(15)00613-0.
Das, C. K., Parekh, A., Parida, P. K., Bhutia, S. K., and Mandal, M. (2019). Lactate dehydrogenase A regulates autophagy and tamoxifen resistance in breast cancer. Biochim. Biophys. Acta Mol. Cell Res. 1866 (6), 1004–1018. doi:10.1016/j.bbamcr.2019.03.004.
Elias, D., Vever, H., Lænkholm, A.-V., Gjerstorff, M. F., Yde, C. W., Lykkesfeldt, A. E., et al. (2015). Gene expression profiling identifies FYN as an important molecule in tamoxifen resistance and a predictor of early recurrence in patients treated with endocrine therapy. Oncogene 34(15), 1919–1927. doi:10.1038/onc.2014.138.
Ferraiuolo, R.-M., Tubman, J., Sinha, I., Hamm, C., and Porter, L. A. (2017). The cyclin-like protein, SPY1, regulates the ERα and ERK1/2 pathways promoting tamoxifen resistance. Oncotarget 8 (14), 23337–23352. doi:10.18632/oncotarget.15578.
Finn, R. S., Crown, J. P., Lang, I., Boer, K., Bondarenko, I. M., Kulyk, S. O., et al. (2015). The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 16 (1), 25–35. doi:10.1016/S1470-2045(14)71159-3.
Gao, A., Sun, T., Ma, G., Cao, J., Hu, Q., Chen, L., et al. (2018). LEM4 confers tamoxifen resistance to breast cancer cells by activating cyclin D-CDK4/6-Rb and ERα pathway. Nat. Commun. 9 (1), 4180. doi:10.1038/s41467-018-06309-8.
Giordano, C., Catalano, S., Panza, S., Vizza, D., Barone, I., Bonofiglio, D., et al. (2011). Farnesoid X receptor inhibits tamoxifen-resistant MCF-7 breast cancer cell growth through downregulation of HER2 expression. Oncogene 30 (39), 4129–4140. doi:10.1038/onc.2011.124.
Gong, C., Man, E. P. S., Tsoi, H., Lee, T. K. W., Lee, P., Ma, S.-T., et al. (2018). BQ323636.1, a novel splice variant to NCOR2, as a predictor for tamoxifen-resistant breast cancer. Clin. Canc. Res. 24 (15), 3681–3691. doi:10.1158/1078-0432.CCR-17-2259.
Gonzalez-Malerva, L., Park, J., Zou, L., Hu, Y., Moradpour, Z., Pearlberg, J., et al. (2011). High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc. Natl. Acad. Sci. USA 108 (5), 2058–2063. doi:10.1073/pnas.1018157108.
Guney Eskiler, G., Cecener, G., Dikmen, G., Egeli, U., and Tunca, B. (2018). Solid lipid nanoparticles: reversal of tamoxifen resistance in breast cancer. Eur. J. Pharmaceut. Sci. 120, 73–88. doi:10.1016/j.ejps.2018.04.040.
Gwak, J. M., Kim, M., Kim, H. J., Jang, M. H., and Park, S. Y. (2017). Expression of embryonal stem cell transcription factors in breast cancer: oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget 8 (22), 36305–36318. doi:10.18632/oncotarget.16750.
He, M., Jin, Q., Chen, C., Liu, Y., Ye, X., Jiang, Y., et al. (2019). The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene 38 (28), 5551–5565. doi:10.1038/s41388-019-0817-3.
Heckler, M. M., Thakor, H., Schafer, C. C., and Riggins, R. B. (2014). ERK/MAPK regulates ERRγ expression, transcriptional activity and receptor-mediated tamoxifen resistance in ER+ breast cancer. FEBS J. 281 (10), 2431–2442. doi:10.1111/febs.12797.
Hortobagyi, G. N., Stemmer, S. M., Burris, H. A., Yap, Y.-S., Sonke, G. S., Paluch-Shimon, S., et al. (2016). Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N. Engl. J. Med. 375 (18), 1738–1748. doi:10.1056/NEJMoa1609709.
Hosford, S. R., and Miller, T. W. (2014). Clinical potential of novel therapeutic targets in breast cancer: CDK4/6, Src, JAK/STAT, PARP, HDAC, and PI3K/AKT/mTOR pathways. Pharmgenomics Pers Med 7, 203–215. doi:10.2147/PGPM.S52762.
Huang, S., Chen, Y., Liang, Z.-M., Li, N.-N., Liu, Y., Zhu, Y., et al. (2019). Targeting Pin1 by all-trans retinoic acid (ATRA) overcomes tamoxifen resistance in breast cancer via multifactorial mechanisms. Front. Cell Dev. Biol. 7, 322. doi:10.3389/fcell.2019.00322.
Ito, T., Kamijo, S., Izumi, H., Kohno, K., Amano, J., and Ito, K.-I. (2012). Alteration of Y-box binding protein-1 expression modifies the response to endocrine therapy in estrogen receptor-positive breast cancer. Breast Canc. Res. Treat. 133 (1), 145–159. doi:10.1007/s10549-011-1731-8.
Jeong, Y., Bae, S. Y., You, D., Jung, S. P., Choi, H. J., Kim, I., et al. (2019). EGFR is a therapeutic target in hormone receptor-positive breast cancer. Cell. Physiol. Biochem. 53 (5), 805–819. doi:10.33594/000000174.
Jeselsohn, R., Cornwell, M., Pun, M., Buchwalter, G., Nguyen, M., Bango, C., et al. (2017). Embryonic transcription factor SOX9 drives breast cancer endocrine resistance. Proc. Natl. Acad. Sci. USA 114 (22), E4482–E4491. doi:10.1073/pnas.1620993114.
Jia, Y., Zhou, J., Luo, X., Chen, M., Chen, Y., Wang, J., et al. (2018). KLF4 overcomes tamoxifen resistance by suppressing MAPK signaling pathway and predicts good prognosis in breast cancer. Cell. Signal. 42, 165–175. doi:10.1016/j.cellsig.2017.09.025.
Jiang, M.-J., Chen, Y.-Y., Dai, J.-J., Gu, D.-N., Mei, Z., Liu, F.-R., et al. (2020). Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Canc. 19 (1), 68. doi:10.1186/s12943-020-01178-6.
Jögi, A., Ehinger, A., Hartman, L., and Alkner, S. (2019). Expression of HIF-1α is related to a poor prognosis and tamoxifen resistance in contralateral breast cancer. PLoS One 14 (12), e0226150. doi:10.1371/journal.pone.0226150.
John, S., Nayvelt, I., Hsu, H.-C., Yang, P., Liu, W., Das, G. M., et al. (2008). Regulation of estrogenic effects by beclin 1 in breast cancer cells. Canc. Res. 68 (19), 7855–7863. doi:10.1158/0008-5472.CAN-07-5875.
Karlsson, E., Veenstra, C., Gårsjö, J., Nordenskjöld, B., Fornander, T., and Stål, O. (2019). PTPN2 deficiency along with activation of nuclear Akt predict endocrine resistance in breast cancer. J. Canc. Res. Clin. Oncol. 145 (3), 599–607. doi:10.1007/s00432-018-2810-6.
Kim, J.-w., and Dang, C. V. (2006). Cancer's molecular sweet tooth and the warburg effect: figure 1. Canc. Res. 66 (18), 8927–8930. doi:10.1158/0008-5472.CAN-06-1501.
Kim, S., Lee, J., Oh, S. J., Nam, S. J., and Lee, J. E. (2015). Differential effect of EGFR inhibitors on tamoxifen-resistant breast cancer cells. Oncol. Rep. 34 (3), 1613–1619. doi:10.3892/or.2015.4116.
Kulkoyluoglu-Cotul, E., Smith, B. P., Wrobel, K., Zhao, Y. C., Chen, K. L. A., Hieronymi, K., et al. (2019). Combined targeting of estrogen receptor alpha and XPO1 prevent akt activation, remodel metabolic pathways and induce autophagy to overcome tamoxifen resistance. Cancers 11 (4), 479. doi:10.3390/cancers11040479.
Kumar, M., Sharma, G., Misra, C., Kumar, R., Singh, B., Katare, O. P., et al. (2018). N-desmethyl tamoxifen and quercetin-loaded multiwalled CNTs: a synergistic approach to overcome MDR in cancer cells. Mater. Sci. Eng. C 89, 274–282. doi:10.1016/j.msec.2018.03.033.
Kuo, S.-J., Lin, H.-Y., Chien, S.-Y., and Chen, D.-R. (2013). SIRT1 suppresses breast cancer growth through downregulation of the Bcl-2 protein. Oncol. Rep. 30 (1), 125–130. doi:10.3892/or.2013.2470.
Lee, M.-H., Koh, D., Na, H., Ka, N.-L., Kim, S., Kim, H.-J., et al. (2018). MTA1 is a novel regulator of autophagy that induces tamoxifen resistance in breast cancer cells. Autophagy 14 (5), 812–824. doi:10.1080/15548627.2017.1388476.
Lee, S., Lee, H., Jeong, D., Ham, J., Park, S., Choi, E. H., et al. (2017). Cold atmospheric plasma restores tamoxifen sensitivity in resistant MCF-7 breast cancer cell. Free Radic. Biol. Med. 110, 280–290. doi:10.1016/j.freeradbiomed.2017.06.017.
Li, D., Ji, H., Niu, X., Yin, L., Wang, Y., Gu, Y., et al. (2020a). Tumor‐associated macrophages secrete CC‐chemokine ligand 2 and induce tamoxifen resistance by activating PI3K/Akt/mTOR in breast cancer. Canc. Sci. 111 (1), 47–58. doi:10.1111/cas.14230.
Li, G., Zhang, J., Xu, Z., and Li, Z. (2020b). ERalpha36 as a potential therapeutic target for tamoxifen-resistant breast cancer cell line through EGFR/ERK signaling pathway. Canc. Manag. Res. 12, 265–275. doi:10.2147/CMAR.S226410.
Li, K., Ying, M., Feng, D., Du, J., Chen, S., Dan, B., et al. (2016). Brachyury promotes tamoxifen resistance in breast cancer by targeting SIRT1. Biomed. Pharmacother. 84, 28–33. doi:10.1016/j.biopha.2016.09.011.
Li, L., Hu, M., Wang, T., Chen, H., and Xu, L. (2019). Repositioning aspirin to treat lung and breast cancers and overcome acquired resistance to targeted therapy. Front Oncol 9, 1503. doi:10.3389/fonc.2019.01503.
Li, W., Xu, L., Che, X., Li, H., Zhang, Y., Song, N., et al. (2018). C-Cbl reverses HER2-mediated tamoxifen resistance in human breast cancer cells. BMC Canc. 18 (1), 507. doi:10.1186/s12885-018-4387-5.
Lin, J. H., Tu, S. H., Chen, L. C., Huang, C. C., Chang, H. L., Cheng, T. C., et al. (2018). Oestrogen receptor-regulated glutathione S-transferase mu 3 expression attenuates hydrogen peroxide-induced cytotoxicity, which confers tamoxifen resistance on breast cancer cells. Breast Canc. Res. Treat. 172 (1), 45–59. doi:10.1007/s10549-018-4897-5.
Liu, J., Yue, W., and Chen, H. (2019). The correlation between autophagy and tamoxifen resistance in breast cancer. Int. J. Clin. Exp. Pathol. 12 (6), 2066–2074.
Liu, T., Liu, P. Y., and Marshall, G. M. (2009). The critical role of the class III histone deacetylase SIRT1 in cancer. Canc. Res. 69 (5), 1702–1705. doi:10.1158/0008-5472.CAN-08-3365.
Lopez-Tarruella, S., and Schiff, R. (2007). The dynamics of estrogen receptor status in breast cancer: re-shaping the paradigm. Clin. Canc. Res. 13 (23), 6921–6925. doi:10.1158/1078-0432.CCR-07-1399.
Lui, A., New, J., Ogony, J., Thomas, S., and Lewis-Wambi, J. (2016). Everolimus downregulates estrogen receptor and induces autophagy in aromatase inhibitor-resistant breast cancer cells. BMC Canc. 16, 487. doi:10.1186/s12885-016-2490-z.
Lv, L., Qimin, W., Xiupeng, L., Lin, S., Huamin, Q., Lifen, W., et al. (2015). Expression and localization of estrogen receptor in human breast cancer and its clinical significance. Cell Biochem. Biophys. 71 (1), 63–68.
Maczis, M. A., Maceyka, M., Waters, M. R., Newton, J., Singh, M., Rigsby, M. F., et al. (2018). Sphingosine kinase 1 activation by estrogen receptor alpha36 contributes to tamoxifen resistance in breast cancer. J. Lipid Res. 59 (12), 2297–2307. doi:10.1194/jlr.M085191.
Mansouri, S., Farahmand, L., Teymourzadeh, A., and Majidzadeh, A. K. (2018a). Clinical evidence on the magnitude of change in growth pathway activity in relation to tamoxifen resistance is required. Curr. Cancer Drug Targets 18 (7), 668–676. doi:10.2174/1568009617666170808110820.
Mansouri, S., Feizi, N., Mahdi, A., Majidzadeh, A. K., and Farahmand, L. (2018b). A review on the role of VEGF in tamoxifen resistance. Anticancer Agents Med. Chem. 18 (14), 2006–2009. doi:10.2174/1871520618666180911142259.
Maqbool, S. N., Lim, S. C., Park, K. C., Hanif, R., Richardson, D. R., Jansson, P. J., et al. (2020). Overcoming tamoxifen resistance in oestrogen receptor-positive breast cancer using the novel thiosemicarbazone anti-cancer agent, DpC. Br. J. Pharmacol. 177 (10), 2365–2380. doi:10.1111/bph.14985.
Maynadier, M., Basile, I., Gallud, A., Gary-Bobo, M., and Garcia, M. (2016). Combination treatment with proteasome inhibitors and antiestrogens has a synergistic effect mediated by p21WAF1 in estrogen receptor-positive breast cancer. Oncol. Rep. 36 (2), 1127–1134. doi:10.3892/or.2016.4873.
Mishra, R. R., Belder, N., Ansari, S. A., Kayhan, M., Bal, H., Raza, U., et al. (2018). Reactivation of cAMP pathway by PDE4D inhibition represents a novel druggable Axis for overcoming tamoxifen resistance in ER-positive breast cancer. Clin. Canc. Res. 24 (8), 1987–2001. doi:10.1158/1078-0432.CCR-17-2776.
Nagelkerke, A., Sieuwerts, A. M., Bussink, J., Sweep, F. C., Look, M. P., Foekens, J. A., et al. (2014). LAMP3 is involved in tamoxifen resistance in breast cancer cells through the modulation of autophagy. Endocr. Relat. Canc. 21 (1), 101–112. doi:10.1530/ERC-13-0183.
Oh, S. J., Kim, O., Lee, J. S., Kim, J. A., Kim, M. R., Choi, H. S., et al. (2010). Inhibition of angiogenesis by quercetin in tamoxifen-resistant breast cancer cells. Food Chem. Toxicol. 48 (11), 3227–3234. doi:10.1016/j.fct.2010.08.028.
Omarjee, S., Jacquemetton, J., Poulard, C., Rochel, N., Dejaegere, A., Chebaro, Y., et al. (2017). The molecular mechanisms underlying the ERalpha-36-mediated signaling in breast cancer. Oncogene 36 (18), 2503–2514. doi:10.1038/onc.2016.415.
Peng, W. X., Huang, J. G., Yang, L., Gong, A. H., and Mo, Y. Y. (2017). Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Canc. 16 (1), 161. doi:10.1186/s12943-017-0727-3.
Putluri, N., Maity, S., Kommagani, R., Creighton, C. J., Putluri, V., Chen, F., et al. (2014). Pathway-centric integrative analysis identifies RRM2 as a prognostic marker in breast cancer associated with poor survival and tamoxifen resistance. Neoplasia 16 (5), 390–402. doi:10.1016/j.neo.2014.05.007.
Qi, H., Jiang, Z., Wang, C., Yang, Y., Li, L., He, H., et al. (2017). Sensitization of tamoxifen-resistant breast cancer cells by Z-ligustilide through inhibiting autophagy and accumulating DNA damages. Oncotarget 8 (17), 29300–29317. doi:10.18632/oncotarget.16832.
Rugo, H. S., Rumble, R. B., Macrae, E., Barton, D. L., Connolly, H. K., Dickler, M. N., et al. (2016). Endocrine therapy for hormone receptor-positive metastatic breast cancer: American society of clinical oncology guideline. J. Clin. Oncol. 34 (25), 3069–3103. doi:10.1200/JCO.2016.67.1487.
Scott, H. S., Antonarakis, S. E., Lalioti, M. D., Rossier, C., Silver, P. A., and Henry, M. F. (1998). Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2). Genomics 48 (3), 330–340. doi:10.1006/geno.1997.5190.
Shen, Y., Zhong, J., Liu, J., Liu, K., Zhao, J., Xu, T., et al. (2018). Protein arginine N-methyltransferase 2 reverses tamoxifen resistance in breast cancer cells through suppression of ER-alpha36. Oncol. Rep. 39 (6), 2604–2612. doi:10.3892/or.2018.6350.
Shi, X. P., Miao, S., Wu, Y., Zhang, W., Zhang, X. F., Ma, H. Z., et al. (2013). Resveratrol sensitizes tamoxifen in antiestrogen-resistant breast cancer cells with epithelial-mesenchymal transition features. Int. J. Mol. Sci. 14 (8), 15655–15668. doi:10.3390/ijms140815655.
Shimoda, M., Hori, A., Wands, J. R., Tsunashima, R., Naoi, Y., Miyake, T., et al. (2017). Endocrine sensitivity of estrogen receptor-positive breast cancer is negatively correlated with aspartate-beta-hydroxylase expression. Canc. Sci. 108 (12), 2454–2461. doi:10.1111/cas.13416.
Sun, W. L., Lan, D., Gan, T. Q., and Cai, Z. W. (2015). Autophagy facilitates multidrug resistance development through inhibition of apoptosis in breast cancer cells. Neoplasma 62 (2), 199–208. doi:10.4149/neo_2015_025.
Thomas, S., Thurn, K. T., Raha, P., Chen, S., and Munster, P. N. (2013). Efficacy of histone deacetylase and estrogen receptor inhibition in breast cancer cells due to concerted down regulation of Akt. PLoS One 8 (7), e68973. doi:10.1371/journal.pone.0068973.
Tong, J. S., Zhang, Q. H., Wang, Z. B., Li, S., Yang, C. R., Fu, X. Q., et al. (2010). ER-alpha36, a novel variant of ER-alpha, mediates estrogen-stimulated proliferation of endometrial carcinoma cells via the PKCdelta/ERK pathway. PLoS One 5 (11), e15408. doi:10.1371/journal.pone.0015408.
Vargo-Gogola, T., and Rosen, J. M. (2007). Modelling breast cancer: one size does not fit all. Nat. Rev. Canc. 7 (9).doi:10.1038/nrc2193.
Vaziri-Gohar, A., Zheng, Y., and Houston, K. D. (2017). IGF-1 receptor modulates FoxO1-mediated tamoxifen response in breast cancer cells. Mol. Canc. Res. 15 (4), 489–497. doi:10.1158/1541-7786.MCR-16-0176.
Viedma-Rodriguez, R., Baiza-Gutman, L., Salamanca-Gomez, F., Diaz-Zaragoza, M., Martinez-Hernandez, G., Ruiz Esparza-Garrido, R., et al. (2014). Mechanisms associated with resistance to tamoxifen in estrogen receptor-positive breast cancer (review). Oncol. Rep. 32 (1), 3–15. doi:10.3892/or.2014.3190.
Wang, B., and Huang, Y. (2020). Effect of aspirin use on neoadjuvant chemoradiotherapy for rectal cancer: a meta-analysis with trial sequential analysis. J. Canc. Res. Clin. Oncol. 146 (3), 2161–2171. doi:10.1007/s00432-020-03222-w.
Wang, H., Tao, L., Qi, K., Zhang, H., Feng, D., Wei, W., et al. (2015). Quercetin reverses tamoxifen resistance in breast cancer cells. J BUON 20 (3), 707–713.
Wang, J., Xie, S., Yang, J., Xiong, H., Jia, Y., Zhou, Y., et al. (2019). The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 12 (1), 81. doi:10.1186/s13045-019-0747-0.
Woo, S. H., Seo, S. K., Park, Y., Kim, E. K., Seong, M. K., Kim, H. A., et al. (2016). Dichloroacetate potentiates tamoxifen-induced cell death in breast cancer cells via downregulation of the epidermal growth factor receptor. Oncotarget 7 (37), 59809–59819. doi:10.18632/oncotarget.10999.
Wu, Y., Yan, B., Xu, W., Guo, L., Wang, Z., Li, G., et al. (2020). Compound C enhances the anticancer effect of aspirin in HER-2-positive breast cancer by regulating lipid metabolism in an AMPK-independent pathway. Int. J. Biol. Sci. 16 (4), 583–597. doi:10.7150/ijbs.39936.
Xue, Y., Lian, W., Zhi, J., Yang, W., Li, Q., Guo, X., et al. (2019). HDAC5-mediated deacetylation and nuclear localisation of SOX9 is critical for tamoxifen resistance in breast cancer. Br. J. Canc. 121 (12), 1039–1049. doi:10.1038/s41416-019-0625-0.
Ye, L., Lin, C., Wang, X., Li, Q., Li, Y., Wang, M., et al. (2019). Epigenetic silencing of SALL2 confers tamoxifen resistance in breast cancer. EMBO Mol. Med. 11 (12), e10638. doi:10.15252/emmm.201910638.
Yin, H., Zhu, Q., Liu, M., Tu, G., Li, Q., Yuan, J., et al. (2017). GPER promotes tamoxifen-resistance in ER+ breast cancer cells by reduced Bim proteins through MAPK/Erk-TRIM2 signaling axis. Int. J. Oncol. 51 (4), 1191–1198. doi:10.3892/ijo.2017.4117.
Yin, L., Zhang, X. T., Bian, X. W., Guo, Y. M., and Wang, Z. Y. (2014). Disruption of the ER-alpha36-EGFR/HER2 positive regulatory loops restores tamoxifen sensitivity in tamoxifen resistance breast cancer cells. PLoS One 9 (9), e107369. doi:10.1371/journal.pone.0107369.
Yu, D., Shi, L., Bu, Y., and Li, W. (2019). Cell division cycle associated 8 is a key regulator of tamoxifen resistance in breast cancer. J Breast Cancer 22(2), 237–247. doi:10.4048/jbc.2019.22.e29.
Zhang, X., and Wang, Z. Y. (2013). Estrogen receptor-alpha variant, ER-alpha36, is involved in tamoxifen resistance and estrogen hypersensitivity. Endocrinology 154 (6), 1990–1998. doi:10.1210/en.2013-1116.
Zhang, Y., Lv, C., Dong, Y., and Yang, Q. (2020). Aspirin-targeted PD-L1 in lung cancer growth inhibition. Thorac Cancer 11(6), 1587–1593. doi:10.1111/1759-7714.13433.
Zhang, Y., Wester, L., He, J., Geiger, T., Moerkens, M., Siddappa, R., et al. (2018). IGF1R signaling drives antiestrogen resistance through PAK2/PIX activation in luminal breast cancer. Oncogene 37 (14), 1869–1884. doi:10.1038/s41388-017-0027-9.
Zhu, Y., Liu, Y., Zhang, C., Chu, J., Wu, Y., Li, Y., et al. (2018). Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat. Commun. 9 (1), 1595. doi:10.1038/s41467-018-03951-0.
Zuo, J. H., Zhu, W., Li, M. Y., Li, X. H., Yi, H., Zeng, G. Q., et al. (2011). Activation of EGFR promotes squamous carcinoma SCC10A cell migration and invasion via inducing EMT-like phenotype change and MMP-9-mediated degradation of E-cadherin. J. Cell. Biochem. 112 (9), 2508–2517. doi:10.1002/jcb.23175.
Keywords: tamoxifen, breast cancer, cell cycle regulators, autophagy, resistance
Citation: Yao J, Deng K, Huang J, Zeng R and Zuo J (2020) Progress in the Understanding of the Mechanism of Tamoxifen Resistance in Breast Cancer. Front. Pharmacol. 11:592912. doi: 10.3389/fphar.2020.592912
Received: 08 August 2020; Accepted: 16 October 2020;
Published: 09 December 2020.
Edited by:
Jin-Ming Yang, University of Kentucky, United StatesReviewed by:
Chia-Che Chang, National Chung Hsing University, TaiwanLoredana Bergandi, University of Turin, Italy
Copyright © 2020 Yao, Deng, Huang, Zeng and Zuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhong Zuo, 632138414@qq.com
†These authors have contributed equally to this work
 Jingwei Yao1,3†
Jingwei Yao1,3† Jianhong Zuo
Jianhong Zuo