Abstract
Objectives
While chest radiograph (CXR) is the first-line imaging investigation in patients with respiratory symptoms, differentiating COVID-19 from other respiratory infections on CXR remains challenging. We developed and validated an AI system for COVID-19 detection on presenting CXR.
Methods
A deep learning model (RadGenX), trained on 168,850 CXRs, was validated on a large international test set of presenting CXRs of symptomatic patients from 9 study sites (US, Italy, and Hong Kong SAR) and 2 public datasets from the US and Europe. Performance was measured by area under the receiver operator characteristic curve (AUC). Bootstrapped simulations were performed to assess performance across a range of potential COVID-19 disease prevalence values (3.33 to 33.3%). Comparison against international radiologists was performed on an independent test set of 852 cases.
Results
RadGenX achieved an AUC of 0.89 on 4-fold cross-validation and an AUC of 0.79 (95%CI 0.78–0.80) on an independent test cohort of 5,894 patients. Delong’s test showed statistical differences in model performance across patients from different regions (p < 0.01), disease severity (p < 0.001), gender (p < 0.001), and age (p = 0.03). Prevalence simulations showed the negative predictive value increases from 86.1% at 33.3% prevalence, to greater than 98.5% at any prevalence below 4.5%. Compared with radiologists, McNemar’s test showed the model has higher sensitivity (p < 0.001) but lower specificity (p < 0.001).
Conclusion
An AI model that predicts COVID-19 infection on CXR in symptomatic patients was validated on a large international cohort providing valuable context on testing and performance expectations for AI systems that perform COVID-19 prediction on CXR.
Key Points
• An AI model developed using CXRs to detect COVID-19 was validated in a large multi-center cohort of 5,894 patients from 9 prospectively recruited sites and 2 public datasets.
• Differences in AI model performance were seen across region, disease severity, gender, and age.
• Prevalence simulations on the international test set demonstrate the model’s NPV is greater than 98.5% at any prevalence below 4.5%.
Similar content being viewed by others
Introduction
With continued uncertainties surrounding vaccine efficacy against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission, waning vaccine efficacy over time, and SARS-CoV-2 variants, early diagnosis will remain central to coronavirus disease 2019 (COVID-19) prevention strategies for the foreseeable future. Timely detection of COVID-19 among symptomatic patients not only reduces transmission, but also allows appropriate treatment and prevention of clinical deterioration [1, 2]. While reverse transcription polymerase chain reaction (RT-PCR) testing remains the gold standard for COVID-19 diagnosis, prolonged test turnaround times, high test and labor costs, and testing reagent shortages have hampered access and use of RT-PCR testing [3, 4]. Indeed, decreasing analytical sensitivity in return for reduced test turnaround time and increased test accessibility can be highly effective at limiting COVID-19 transmission [5]. Accessible, accurate, and cost-effective COVID-19 screening tools are urgently needed so that symptomatic patients can be better triaged and confirmatory RT-PCR testing can be preferentially performed in those with a high pre-test probability of having and being likely to transmit COVID-19.
While computed tomography (CT) imaging has been used for both COVID-19 detection and disease characterization [6], it is clinically not indicated for the majority of patients with mild disease. Furthermore, its high financial costs and relative low throughput effectively limit its utility to first world nations. Comparatively, a chest radiograph (CXR) is often routinely obtained for the initial assessment of patients with acute respiratory symptoms who seek medical care, is cheaper, and has lower radiation dose, and it is available in clinics worldwide [7]. Thus, CXR has the potential to be used to aid COVID-19 detection in symptomatic patients without requiring additional resources. Unfortunately, detecting COVID-19 on CXR is difficult as a significant proportion of patients lack characteristic CXR findings that can be used to differentiate it from other respiratory infections [8].
Recent studies have shown that artificial intelligence (AI) models employing computer vision–based deep neural networks can detect and learn COVID-19 features on CXR that may be nonobvious to radiologists [9,10,11,12,13]. While promising, many AI models suffer from training dataset bias and poor generalizability [14]. Moreover with many studies focusing on detecting COVID-19 from healthy individuals, they do not address the more clinically relevant question of differentiating COVID-19 from other causes in patients with respiratory symptoms; hence, the true performance of these AI models in a clinically relevant setting remains unknown [15]. To address these challenges, we conducted a large international validation study of a COVID-19 CXR AI prediction model (RadGenX) on symptomatic patients suspected to have COVID-19 [16, 17].
Methods
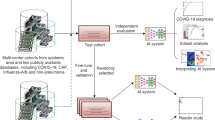
This was a retrospective, observational multi-center study whose primary objective was to evaluate RadGenX’s performance on predicting COVID-19 from CXR in symptomatic COVID-19 suspected patients. The overall study design is shown in Fig. 1.
International test set
Independent external testing was performed on a combined cohort consisting of CXRs of patients collected from: (1) 9 independent sites involving 11 hospitals (hereafter referred as “private” study patients) across the United States (US) (N = 4), Italy (N = 5), and Hong Kong SAR, China (N = 2), AND (2) two large public databases (hereafter referred as “public” study patients): one from Europe and the other from the US [18, 19]. The “international test set” consisted of the 9 private study sites plus 1 of the “public” sites (COVID-19-NY-SBU – see below). All study patients were patients ≥ 18 years old that presented with respiratory symptoms and underwent CXR and SARS-CoV-2 RT-PCR testing. The study was approved by the Institutional Review Board of each participating site and followed the guidelines outlined in the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) in the reporting of the study [20].
Private study patients
Per study protocol, private study patients were collected between February 1, 2020, and July 30, 2020; all study patients must have had respiratory symptoms, including but not exclusive to cough, dyspnea, or signs and symptoms of a respiratory tract infection. They must have also undergone SARS-CoV-2 RT-PCR testing with a definitive positive or negative result; patients who had equivocal or conflicting results were excluded. Historic negatives could be included by the study sites, which was defined as pre-COVID-19 era patients who similarly presented to their respective study site with respiratory symptoms prior to August 1, 2019. Only frontal CXRs were included. Demographic and clinical data (including symptom severity) of patients at presentation were also collected. Patient symptoms were stratified using the 9-point ordinal scale as recommended by the World Health Organization in COVID-19 studies into 3 categories: (mild = 1–2, moderate = 3–4, and severe = 5–7) [21]. Any patients with scores of either 0 (asymptomatic/uninfected) or 8 (dead) were excluded per study protocol.
Public study patients
In addition, a comprehensive search was performed (December 1, 2021) to identify additional public datasets for external validation with greater than 500 patients per set meeting similar criteria [22]. Of the publicly available datasets, two met these criteria and did not overlap with our training cohort. COVIDGR contained 852 COVID-19 positive and negative cases from Granada, Spain [18]. COVID-19-NY-SBU included 1,384 COVID-19 positive cases acquired at Stony Brook University [19]. Cases from the two public datasets similarly included in (addition to digital CXR images) basic demographics (i.e., age and gender), definitive COVID-19 status, and disease severity.
Training RadGenX AI model using a centralized federated learning system
RadGenX is based on the previously reported RadGen CXR AI prediction model [16, 17]. This original RadGen model was then updated using data from private study patients and 2 public SARS-CoV-2 CXR datasets (see supplement). Conventional computer vision–based medical AI models typically mandate centralization of data which raises concerns for patient privacy, while classical weight-based federated models require significant compute and technical resources not readily available to the vast majority of imaging centers [23]. In order to (1) allow sites with minimal compute resources or technical knowledge to collaboratively train a shared AI model, and (2) enable these sites to still maintain sole custodianship of their patient data, we modified RadGen into a standalone Windows PC compatible software application and enabled privacy-preserving data sharing for model updating using a modified federated learning approach [24].
Of the 9 study sites, 3 sites contributed 3,650 CXRs of 2,576 patients (43.1% COVID-19 positive) between February 1, 2020, and July 30, 2020, for model updating (Table S1) (Fondazione Policlinico Universitario Agostino Gemelli IRCCS – Università Cattolica del Sacro Cuore Roma, Fondazione I.R.C.C.S. Policlinico San Matteo and University of Brescia).
The public datasets used for model updating (accessed September 15, 2020) consisted of a total of 18,072 CXRs (11,953 positive and 6,119 negative) from the BIMCIV-COVID-19 and 252 positive CXRs from the COVID-19-AR datasets [25, 26].
RadGenX model training and validation
The architectures of both RadGen and RadGenX are based on SE-ResNeXt-50-32x4d [24]. RadGenX was trained using the SGD optimizer for 120 epochs, with an initial learning rate of 0.01, decreased by 1/5 at epochs 70, 90, 110 and with norm gradient clipping of 8.0. Hard negative mining was also performed [23], by applying the partially trained model to the CheXpert dataset with the top 2,000 CXRs with the highest probability scores then considered in the loss function [27]. To offset positive-to-negative case imbalance, we generated 4 separate models with each model separately trained on each of the 4 non-overlapping data folds.
For model validation, we performed 4-fold cross-validation with each of the 4 separate models separately trained and validated on each of the 4 non-overlapping folds. A threshold based on obtaining 90.0% sensitivity was selected a priori (based on published guidelines recommended by regulatory bodies for minimum performance for serological tests for SARS-CoV-2 detection), which was applied to the aggregated held out validation sets for each of the 4 models in order to identify the corresponding cut point [28]. The final model, RadGenX, consisted of an equally weighted ensemble of the 4 models whose output was a binary prediction (based on application of the pre-specified cut-point) of SARS-CoV-2 RT-PCR positivity for a given input DICOM frontal CXR. This model was independently evaluated on the international test set.
COVID-19 prevalence analysis
While the case-control independent test set allows broad validation of RadGenX, in order to assess the performance of RadGenX under varying COVID-19 prevalences, we additionally ran a series of bootstrapped prevalence simulations leveraging data from the 9 study sites from the international test set (n = 4,559) across simulated disease prevalences in step-wise fashion from 3.33% (1:30) to 33.3% (1:2). The performance of RadGenX was reported as the mean prediction score at each of the simulated prevalence levels, based on a random sampling from the international test set of 100 COVID-19 positive patients against the corresponding number of randomly selected COVID-19 negative patients to meet each prevalence ratio, bootstrapped 10,000 times.
RadGenX versus reader study
In order to better understand differences in performance between RadGenX and radiologists, we performed a head-to-head comparison analysis on the COVIDGR dataset [18]. Three radiologists from the US, Italy, and HK, all board-certified, and each with over 10 years’ experience (KWHC, DSW, ARL) analyzed all 852 CXRs in the COVIDGR dataset (Table S2). The readers were given the CXRs in a blinded and random order which each reader independently reviewed. Assessment of each CXR image consisted of evaluating the presence or absence of findings consistent with COVID-19 pneumonia. Reader consensus was created using the majority vote method for a given image for each of these two tasks. Their predictions were compared to those of RadGenX and the individual patient ground truth COVID-19 status.
Statistical analysis
The performances of RadGenX and the radiologists were evaluated using AUC, sensitivity, and specificity. In subgroup analysis, odds ratios were calculated and Wilcoxon test was used to compare subgroups with the overall performance of RadGenX. In the reader study, interclass correlation coefficient (ICC, two-way mixed, average score) values and 95% CI were used to assess inter-rater reliability. ICC values of 0–0.25, 0.26–0.49, 0.50–0.69, 0.70–0.89, and 0.90–1.00 indicated little or no reliability, low reliability, moderate reliability, high reliability, and very high reliability, respectively. Pearson’s chi-squared tests were employed to compare RadGenX and radiologists. DeLong test was used to subgroup model performances. A p value < 0.05 was considered statistically significant. Analyses were performed with the use of R software, version 3.6.3 (R Foundation for Statistical Computing).
Results
RadGenX achieved an area under the curve (AUC) of 0.89 (95% confidence interval [CI] 0.88–0.89) across the 4 individual models on 4-fold cross-validation (Fig. 2A). The corresponding specificity at the pre-specified cut point selected to achieve 90% COVID-19 test sensitivity (see “Methods”) was 62.6% (Fig. 2B).
Evaluation of RadGenX on the international test set
Of the 8,380 symptomatic COVID-19 suspected cases curated by our 9 private studies sites, a combined 2,202 patients were used for model updating. Of the remaining 6,178 patients, 1,619 did not meet the study eligibility criteria, resulting in a total of 4,559 patients included in the international test set. This cohort consisted of 1,336, 1,456, and 1,767 patients from the US, Italy, and HK, respectively. For each country, the percentage of RT-PCR positive COVID-19 patients was 28.3%, 42.2%, and 23.8% for the US, Italy, and HK, respectively.
The NY-SBU dataset, consisting of 1,335/1,384 (100% COVID-19 positive) (mean age 57.5 [range 18 to 90], 56.5% men) that met inclusion criteria was also included in the international test set. For subgroup analysis, cases from NY-SBU was grouped with the private US cohorts. The final demographics of the international test set are summarized in Table 1 (Tables S3 & S4).
On this composite independent test set of 5,894 diverse patients from both the privately and publicly curated datasets, RadGenX achieved an overall AUC of 0.79 (95% CI 0.78 to 0.80) for detecting RT-PCR-confirmed COVID-19 on presenting CXR on symptomatic COVID-19 suspected patients (Fig. 3A). The overall sensitivity and specificity of the model were 79.1% (95% CI 77.6 to 80.6%) and 60.5% (95% CI 58.8 to 62.2%), respectively. There was no significant difference in model performance when comparing RT-PCR negatives versus historical negatives as controls (AUC = 0.80 [95% CI 0.78 to 0.81] vs 0.81 [95% CI 0.80 to 0.83]; p = 0.11).
RadGenX performance on the international test set. A ROC curve of RadGenX on the international test set (N = 5,894); AUC = 0.79 (95%CI 0.78–0.81). B RadGenX’s AUC with 95% confidence intervals (CI) across different regions, disease severity, gender and age; C CXR of a 60-year old male COVID-19 positive patient with mild symptoms at the time of diagnosis showing minimal air space opacities in right lower zone (yellow arrow) that RadGenX correctly predicted as COVID-19 positive. D A 67-year old male COVID-19 negative patient with severe symptoms at presentation showing extensive consolidation in both lungs (yellow arrows) on chest X-ray that RadGenX called COVID-19 negative. E CXR of a 49-year-old male with mild respiratory symptoms for 2–3 days with a normal appearing chest radiograph that RadGenX correctly predicted as COVID-19 positive
Reflective of the diverse patient composition, this cohort was similarly diverse in disease severity, region, age, and gender. We therefore explored whether these factors affected model performance. As expected, there were model performance differences as assessed by AUC across disease severity (0.72 vs 0.81 vs 0.90, for mild, moderate, and severe disease respectively, p < 0.001 for any combination), region (0.71 vs 0.85 vs 0.78 for US, Europe, and Hong Kong respectively, p < 0.01 for any combination), gender (0.82 vs 0.75 between male and female, p < 0.001), and age (0.78 vs 0.80 for < 65 and ≥ 65 years, p = 0.03) (Fig. 3B–E).
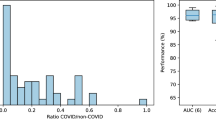
While a test’s sensitivity and specificity are fixed, variable prevalence analysis over 10,000 simulations performed on the international test set, revealed that the model’s positive predictive value (PPV) steadily increased from 8.8% at 3.33% prevalence to 50.5% at 33.3%. Conversely, the negative predictive value (NPV) increased from 86.1% at 33.3% prevalence to greater than 98.5% at any COVID-19 prevalence below 4.5%.
RadGenX compared to radiologists
The COVIDGR public dataset consisted of 852 patients (50% COVID-19 positive) and was used as the external test set for the radiologist-AI comparison study. The radiologists’ agreement overall was high with an interclass correlation (ICC) of 0.81 (95% CI 0.78 to 0.83) for detecting COVID-19. The overall sensitivity and specificity of the radiologists for detecting COVID-19 on CXR were 51.6% (95% CI 46.9 to 56.4%) and 99.1% (95% CI 97.6 to 99.6%), respectively. RadGenX achieved an overall AUC of 0.82 (95% CI 0.79 to 0.85). Applying the pre-specified cut-point, the sensitivity and specificity of the model were 82.9% (95% CI 79.0 to 86.1%) and 56.8% (95% CI 52.1 to 61.4%) respectively (Fig. 4). Radiologists and RadGenX had false positive rates of 1% (4/426), and 43.2% (184/426), respectively, and false negative rates of 48.4% (206/426) and 17.1% (73/426), respectively.
RadGenX performance compared to radiologists. Receiver operator characteristic (ROC) curve of RadGenX on the independent COVIDGR dataset (n = 852) with the performance of the 3 board-certified radiologists from the US, Italy, and Hong Kong SAR each denoted on the plot [18]
Discussion
In summary, we validated the performance of an AI model in detecting COVID-19 on CXR in patients presenting with respiratory symptoms in a cohort of 5,894 patients from 3 different continents. The model achieved an AUC of 0.79, sensitivity of 79.1%, and specificity of 60.5%. The size and diversity of our test set allowed us to evaluate the model’s “true” performance across a broad cross section of age groups, disease conditions, localities, and healthcare systems, which is particularly relevant among growing concerns of “brittle” AI, model performance overestimations, and cohort bias [29, 30]. Further, prevalence simulations demonstrated the model’s performance profile across a range of potential COVID-19 prevalence levels, revealing that the model’s NPV steadily increased from 86.1% at 33.3% prevalence, and exceeded 98.5% at any COVID-19 level below 4.5%. Finally, comparison of RadGenX against radiologists from Asia, the US, and EU, on an independent set of 852 positive and negative COVID-19 cases, resulted in an AUC of 0.82, sensitivity of 82.9%, and specificity of 56.8%, compared to a sensitivity and specificity of 51.6% and 99.1%, respectively, for the radiologists. The scale and scope of this study provides valuable context and perspective on realistic performance expectations for AI systems that perform COVID-19 prediction on CXR and, more broadly, on the challenges of achieving truly “generalizable” diagnostic AI models, even when the ground truth is an objective standard such as PCR testing.
Our international test set consisted of a large and geographically diverse representation of COVID-19 cases consisting of cases collected from 10 different institutions across 3 different continents. Unlike many studies that assessed AI model performance in differentiating symptomatic COVID-19 patients from normal healthy individuals, we specifically evaluated our model’s ability to differentiate among patients with respiratory symptoms on their presenting CXR [15]. We believe this is a more impactful and less well studied task as the clinically relevant conundrum arises when a symptomatic patient is first admitted to hospital as indiscriminate COVID-19 mass screening, regardless of technology, has so far shown to be expensive and of unclear benefit [31]. The more even distribution of mild, moderate, and severe hospitalized COVID-19 cases we observed in our COVID-19 positive group compared to the negative group, is likely a reflection of the variation in country-by-country COVID-19 management strategies witnessed during the early phases of the COVID-19 pandemic. For example, a number of countries in Asia initially pursued a stringent containment policy whereby all PCR positive cases were hospitalized, regardless of disease severity, whereas many US and European systems tended to hospitalize only sick COVID positive patients, resulting in a relative enrichment of mild cases in the COVID-19 positive group (26.3%) compared to the negative group (4.8%). The resulting relatively even distribution in disease severity across mild, moderate, and severe in the positive group, powered by the large total number of patients studied, fortunately provided a unique opportunity to also more fully understand the impact of disease severity on model performance as we have highlighted in the results.
Given the size and diversity of our international test set, it is not surprising that differences in performance on subgroup analysis were observed [32]. Although the model performed equally well across both the privately curated and public datasets and the historical cohort and RT-PCR proven cases, unsurprisingly, significant performance differences were observed between age groups, gender, locality, and disease severity. In addition to providing insights into specific factors that increase or decrease model performance across this large study population, this also highlights one of the difficulties encountered when attempting to train a sufficiently generalizable model that often requires balancing capturing a large number of diverse cases with patient-specific data which may be difficult to balance due to patient privacy concerns. In our specific instance, it remains to be seen whether training with even larger datasets and more balanced annotated cohorts would be sufficient to overcome these subgroup differences or whether these are inherent limitations of the current model, or of the larger set of diagnostic AI models aiming to be broadly “generalizable,” or whether it is simply a feature of this particular disease entity.
An important study limitation is whether the ongoing emergence of SAR-CoV-2 variants will affect the performance of the current RadGenX model. A number of these variants have shown increased transmissibility, morbidity, and mortality [33]. Although initial data suggest that their radiographic appearance is similar to that of the original strain, it remains unknown whether these variants will lead to a substantial divergence in radiographic phenotypes. As we have similarly demonstrated the critical importance of training set diversity on model generalizability, it will be imperative to continue to capture CXR data from SAR-CoV-2 variants as COVID-19 infections continue to surge globally. The centralized federated learning framework we employed thus enables model updating in a privacy-preserving manner allowing for incorporation of new SAR-CoV-2 variant CXR data into future iterations of our model.
In this study, we used SARS-CoV-2 RT-PCR test results as the ground truth reference for COVID-19 infection [34]. While the implication is that the performance of our AI model will never surpass nor replace RT-PCR testing, this is clearly not the model’s intent, any more so than low-dose lung CT or virtual colonoscopy is meant to replace tissue diagnosis; instead, the goal of tools such as RadGenX is to serve as an adjunct to COVID-19 RT-PCR testing [5]. Tests such as CXR are more accessible and offer faster turnaround time, and not only have the potential to improve cost effectiveness and PCR utilization, but also have been shown to have clear benefits for infection containment, which is particularly relevant in resource-constrained regions [35].
This is supported by our prevalence analysis where we interrogated the model in stepwise fashion across a range of simulated COVID-19 prevalence levels, representing on one end a “surge” prevalence of 33% as recently seen in Hong Kong and other nations [36], sequentially down to reported “endemic” prevalence rates near 3% [37]. Given the model’s excellent NPV—exceeding 98.5% at any prevalence below 4.5%—a potential application could be as an adjunctive screening tool to PCR in symptomatic patients during periods when prevalence nears endemic levels, where a negative test result could be used to effectively exclude patients from PCR testing with high confidence, thus reserving PCR testing for those with higher likelihood to be PCR positive. This could potentially offer a cost-effective option for COVID-19 testing in resource-constrained regions. Prospective studies will ultimately be needed to validate its role in such a scenario.
In summary, we have validated an AI model that predicts COVID-19 status from CXR in symptomatic patients in one of the largest and most diverse, independent study populations to date. We also provide modeling analysis to show how the model performs under a wide range of simulated COVID-19 disease prevalence levels and compare it to radiologists, highlighting its potential role as an adjunctive diagnostic tool to PCR testing with good sensitivity and high NPV, which may be of utility in resource-constrained regions when COVID-19 levels are endemic. Additionally, implementation of RadGenX is simple, requiring only digital radiography and a personal computer—both of which are ubiquitous in medical clinics worldwide. More broadly, this study provides a set of parameters and realistic expectations that could be useful for studying and gauging radiological AI model performance, which is particularly relevant as an increasing number of AI models seek broad and general deployment across diagnostic imaging practices.
Abbreviations
- AI:
-
Artificial intelligence
- CXR:
-
Chest X-ray
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
References
Gottlieb RL, Vaca CE, Paredes R et al (2021) Early remdesivir to prevent progression to severe Covid-19 in outpatients. N Engl J Med. https://doi.org/10.1056/NEJMoa2116846
Kucharski AJ, Klepac P, Conlan AJK et al (2020) Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect Dis 20:1151–1160
Dryden-Peterson S, Velásquez GE, Stopka TJ, Davey S, Lockman S, Ojikutu BO (2021) Disparities in SARS-CoV-2 testing in Massachusetts during the COVID-19 pandemic. JAMA Netw Open 4:e2037067
Quilty BJ, Clifford S, Hellewell J et al (2021) Quarantine and testing strategies in contact tracing for SARS-CoV-2: a modelling study. Lancet Public Health 6:e175–e183
Mina MJ, Parker R, Larremore DB (2020) Rethinking Covid-19 test sensitivity — a strategy for containment. N Engl J Med 383:e120
Fang Y, Zhang H, Xie J et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 296:E115–E117
Woodhead M, Blasi F, Ewig S et al (2011) Guidelines for the management of adult lower respiratory tract infections--full version. Clin Microbiol Infect 17(Suppl 6):E1–E59
Rubin GD, Ryerson CJ, Haramati LB et al (2020) The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology 296:172–180
Wang L, Lin ZQ, Wong A (2020) COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep 10:19549
Wehbe RM, Sheng J, Dutta S et al (2021) DeepCOVID-XR: an artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large U.S. clinical data set. Radiology 299:E167–E176
Oh Y, Park S, Ye JC (2020) Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans Med Imaging 39:2688–2700
Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi A (2020) COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit Lett 138:638–643
Sharma A, Rani S, Gupta D (2020) Artificial intelligence-based classification of chest X-ray images into COVID-19 and other infectious diseases. Int J Biomed Imaging 2020:8889023
Dhont J, Wolfs C, Verhaegen F (2021) Automatic COVID-19 diagnosis based on chest radiography and deep learning – success story or dataset bias? Med Phys. https://doi.org/10.1002/mp.15419
Gillman AG, Lunardo F, Prinable J et al (2021) Automated COVID-19 diagnosis and prognosis with medical imaging and who is publishing: a systematic review. Phys Eng Sci Med. https://doi.org/10.1007/s13246-021-01093-0
Chiu WHK, Vardhanabhuti V, Poplavskiy D et al (2020) Detection of COVID-19 using deep learning algorithms on chest radiographs. J Thorac Imaging 35:369–376
Chiu WHK, Poplavskiy D, Zhang S, Ho Yu PL, Kuo MD (2021) Dynamic Prediction of SARS-CoV-2 RT-PCR status on Chest Radiographs using Deep Learning Enabled Radiogenomics. medRxiv. https://doi.org/10.1101/2021.01.10.21249370
Tabik S, Gomez-Rios A, Martin-Rodriguez JL et al (2020) COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest X-ray images. IEEE J Biomed Health Inform 24:3595–3605
Saltz J, Saltz M, Prasanna P et al (2021) Stony Brook University COVID-19 Positive Cases [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.BBAG-2923
Mongan J, Moy L, Kahn CE Jr (2020) Checklist for Artificial Intelligence in Medical Imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell 2:e200029
Blueprint WJG, Switzerland (2020) novel Coronavirus, COVID-19 Therapeutic Trial Synopsis.
Garcia Santa Cruz B, Bossa MN, Sölter J, Husch AD (2021) Public Covid-19 X-ray datasets and their impact on model bias – a systematic review of a significant problem. Med Image Anal 74:102225
Zhao Y, Li M, Lai L, Suda N, Civin D, Chandra V (2018) Federated learning with non-iid data. arXiv preprint arXiv:180600582
Pianykh OS, Langs G, Dewey M et al (2020) Continuous learning AI in radiology: implementation principles and early applications. Radiology 297:6–14
Iglesia-Vayá Mdl, Saborit JM, Montell JA et al (2020) BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients. abs/2006.01174
Desai S, Baghal A, Wongsurawat T et al (2020) Chest imaging representing a COVID-19 positive rural U.S. population. Sci Data 7:414
Bustos A, Pertusa A, Salinas J-M, de la Iglesia-Vayá M (2019) PadChest: a large chest x-ray image dataset with multi-label annotated reports. arXiv e-prints
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer, pp 234–241
Roberts M, Driggs D, Thorpe M et al (2021) Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell 3:199–217
Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK (2018) Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med 15:e1002683
Cao S, Gan Y, Wang C et al (2020) Post-lockdown SARS-CoV-2 nucleic acid screening in nearly ten million residents of Wuhan, China. Nat Commun 11:5917
Goel K, Gu A, Li Y, Ré C (2020) Model patching: closing the subgroup performance gap with data augmentation. arXiv preprint arXiv:200806775
Kirby T (2021) New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir Med 9:e20–e21
Zhang Z, Bi Q, Fang S et al (2021) Insight into the practical performance of RT-PCR testing for SARS-CoV-2 using serological data: a cohort study. Lancet Microbe 2:e79–e87
Larremore DB, Wilder B, Lester E et al (2021) Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv 7:eabd5393
The University of Hong Kong (2022) HKUMed proposes forward planning after Hong Kong’s fifth wave of Omicron BA. https://sph.hku.hk/en/News-And-Events/Press-Releases/2022/HKUMed-proposes-forward-planning-after-Hong-Kong
Yiannoutsos CT, Halverson PK, Menachemi N (2021) Bayesian estimation of SARS-CoV-2 prevalence in Indiana by random testing. Proc Natl Acad Sci U S A 118
Acknowledgements
The following authors have also contributed to the manuscript:
Andrea Leonardi (MD)1, Carlo Catalano (MD)1, Paolo Ricci (MD)1, Hiu Yin S. Lam (MBBS)2, Ho Yuen F. Wong (MBBS)2, Gilbert Lui (PhD)3, Nicoletta Izzi1 (MD)4, Antonella Donatelli (MD)4, Francesca Marchetti (MD)4, Annie Rhee (MD)5, Lorenzo Preda (MD)6, Nicola Carapella (MD)7, Helen Zhi (PhD)8, Francesco Ascari (MD)9,10, Patrizia Lazzari (MD)9,10, Leonardo Canulli (MD)9,11, Pietro Torricelli (MD)9,10, San Ming P. Yu (MBBS)12, Yu Wai T. Hon (MBBS)12, Yee Hing J. Hui (MBBS)12, Cesare Colosimo (MD)13,14, Luigi Natale (MD)13,14, Riccardo Marano (MD)13,14, Maurizio Sanguinetti (MD)13,15
Affiliations:
1Department of Radiological, Oncological and Pathological Sciences, Sapienza University of Rome, Rome; Italy
2Radiology Department, Queen Mary Hospital; Hong Kong SAR, China
3Department of Medicine, LKS Faculty of Medicine, The University of Hong Kong; Hong Kong SAR, China
4Department of Clinical, Surgical, Diagnostic and Pediatric Sciences, University of Pavia; Italy
5Radiology Department, Mayo Clinic; Rochester, Minnesota, USA
6Department of Radiology, Fondazione IRCCS Policlinico San Matteo; Italy
7Department of Medical and Surgical Specialties, Radiological Sciences and Public Health, University of Brescia, ASST Spedali Civili of Brescia; Italy
8School of Public Health, LKS Faculty of Medicine, The University of Hong Kong; Hong Kong SAR, China
9Department of Medical and Surgical Sciences for Children & Adults, Modena and Reggio Emilia University; Italy
10Division of Radiology, Azienda Ospedaliero-Universitaria Policlinico di Modena; Italy
11Division of Education, Research and Innovation, Azienda Ospedaliero-Universitaria Policlinico di Modena; Italy
12Radiology Department, United Christian Hospital; Hong Kong SAR, China
13Department of Radiological and Hematological Sciences, Section of Radiology, Università Cattolica del Sacro Cuore; Rome, Italy
14Department of Diagnostic Imaging, Oncological Radiotherapy and Hematology, Fondazione Policlinico Universitario “A. Gemelli” IRCCS; Rome, Italy
15Institute of Microbiology and Division of Clinical Microbiology, Università Cattolica del Sacro Cuore, Rome; Italy
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantors of this publication are MDK and KWHC.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: MDK is a founder, shareholder and unpaid part-time employee of Ensemble Group. DP is a part-time employee of Ensemble Group. The remaining authors declare that they have no competing interests. The remaining authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
HZ kindly provided statistical advice for this manuscript and has significant statistical expertise. However, no complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board for each participating institution.
Ethical approval
Institutional Review Board approval was obtained for each participating institution.
Methodology
• Retrospective
• Observational
• Multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 98 kb)
Rights and permissions
About this article
Cite this article
Kuo, M.D., Chiu, K.W.H., Wang, D.S. et al. Multi-center validation of an artificial intelligence system for detection of COVID-19 on chest radiographs in symptomatic patients. Eur Radiol 33, 23–33 (2023). https://doi.org/10.1007/s00330-022-08969-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08969-z