Abstract
SRC-3/AIB1 (steroid receptor coactivator 3/amplified in breast cancer 1) is an authentic oncogene that contributes to the development of drug resistance and poor disease-free survival in cancer patients. Autophagy is also an important cell death mechanism that has tumor suppressor function. In this study, we identified macrophage migration inhibitory factor (MIF) as a novel target gene of SRC-3 and demonstrated its importance in cell survival. Specifically, we showed that MIF is a strong suppressor of autophagic cell death. We further showed that suppression of MIF, in turn, induced autophagic cell death, enhanced chemosensitivity and inhibited tumorigenesis in a xenograft mouse tumorigenesis model. Our study demonstrated that regulation of MIF expression and suppression of autophagic cell death is a potent mechanism by which SRC-3 contributes to increased chemoresistance and tumorigenicity.
Similar content being viewed by others
Introduction
Breast cancer is the most common non-dermal malignancy and the most common cause of cancer-related deaths in women. Even with early detection and improved treatments, the 5-year survival rate in patients with advanced breast cancer is about 20% 1. Resistance to therapy plays a major role in treatment failure. To improve treatment outcome, a better understanding of the molecular mechanisms underlying such drug resistance is critical.
Although the exact cause of breast cancer and drug resistance is not known, both genetic and non-genetic factors are involved. Steroid receptor coactivator 3 (SRC-3/AIB1/ACTR/RAC3/p/CIP), a member of the p160 family of coactivators, is an oncogene that interacts with nuclear receptors (NRs) and non-NR transcriptional factors, including estrogen receptor (ER), progesterone receptor (PR), androgen receptor and NF-κB and is required for the expression of subsets of their respective target genes. By modulating gene expression, SRC-3 regulates diverse physiological functions and has been implicated in the development of breast cancer and prostate cancer. SRC-3 (also known as AIB1 for amplified in breast cancer 1) was initially identified as a gene amplified and overexpressed in 5%-10% of ovarian cancers and 30%-60% of breast cancers 2. Recent clinical evidence showed that high levels of SRC-3 expression were associated with high levels of HER2/neu, EGFR, development of drug resistance and poor disease-free survival in patients with breast and lung cancers 3,4,5. These results suggest that crosstalk between signaling pathways regulate SRC-3 activity through protein post-translational modifications (PTMs) 6,7,8. These dynamic and reversible PTMs link SRC-3 functions and cellular response to extracellular stimuli and underscore another important aspect of the oncogenic function of SRC-3.
Despite its discovery in the mid-1960s as a T-cell-derived factor that modulated the motility of monocytes, macrophage migration inhibitory factor (MIF) has been shown to be involved in cancers 9,10,11,12,13. Interestingly, both circulating and intracellular levels of MIF were elevated in patients with cancers, and the levels of MIF were closely correlated with tumor aggressiveness and metastatic potential, suggesting that MIF contributed to disease severity and survival 14,15,16,17. Intracellular MIF interacted with the signalosome component JAB1/CSN5 and was shown to regulate both the activity of tumor suppressor p53 and the angiogenesis induced by hypoxia 17, 18. Interestingly, JAB1/CSN5 was further shown to interact with PR and SRC-1 and potentiated the activity of a variety of transcription factors known to associate with SRC-1, such as AP-1 and NF-κB 19,20,21. In addition, JAB1 expression was required for the rapid and transient activation of ERK by MIF, and the ability of JAB1/CSN5 to activate AP-1 is modulated by interaction with MIF 22, 23. The extracellular action of MIF was mediated through binding of its receptors on target cells, including MHC class II chaperone CD74 and chemokine receptors CXCR2 and CXCR4, followed by activation of downstream signaling pathways 10, 24, 25. Together, these findings clearly suggested a potential involvement of MIF in cancer, but the precise role(s) of MIF remains to be elucidated.
Autophagy (type II programmed cell death) is a highly regulated process that is involved in numerous physiological functions in multicellular organisms, including organelle turnover and protein degradation 26, 27. Although some evidence suggested that autophagy promotes cell survival under nutrient deprivation, a growing body of evidence suggested that suppression of autophagic cell death promoted cancer development. The major evidence that supported the tumor suppressive function of autophagy includes the following: (1) monoallelic loss of the essential autophagy gene beclin 1 was found with high frequency in human breast, ovarian and prostate tumors; (2) an increase in tumor incidence was observed in beclin 1+/− mutant mice; and (3) DAP-K and PTEN tumor suppressors upregulate autophagic pathways 28,29,30,31,32. Furthermore, the activity of autophagy is connected to apoptosis. Depending on the cellular context and stimuli, in some instances, autophagy precedes apoptosis and may enhance or inhibit apoptosis, while in others, autophagy and apoptosis are mutually exclusive and function as backups for each other to ensure complete cell death. Although the relationship between autophagy and apoptosis is not well understood, the anti-apoptotic Bcl-2 family proteins have been shown to serve as a switch between these two cell death mechanisms 33. Since Bcl-2 is an oncogene that is overexpressed in 50%-70% of breast cancers, inhibition of autophagic and apoptotic cell death by Bcl-2 may promote resistance to chemotherapy and hormone therapies 34,35,36.
In this report, we demonstrate that SRC-3 regulates the expression of MIF and show that suppression of SRC-3 or MIF expression reduces cell viability through induction of autophagic cell death in MCF-7 breast cancer cells. Importantly, suppression of MIF increased autophagic cell death and is associated with reduced tumorigenicity and enhanced chemosensitivity. Together, our findings support a role of autophagy in tumor suppression and suggest that induction of autophagic cell death by suppression of SRC-3 or MIF is a novel anticancer mechanism.
Results
SRC-3 regulates MIF expression
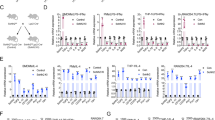
Recent reports indicate that overexpression of oncogene SRC-3 was associated with development of drug resistance and poor disease-free survival in patients with breast cancer 4, 5. To better understand the mechanism by which SRC-3 promotes drug resistance and cell survival, we devised an assay to identify genes that can promote cell proliferation and cell survival when expression of SRC-3 was suppressed in breast cancer cells. Infection of MCF-7 breast cancer cells with lentivirus expressing short hairpin RNA (shRNA) against SRC-3 resulted in a significant (∼98%) reduction of SRC-3 expression, but did not affect the levels of SRC-1 or SRC-2 (Figure 1A). Knockdown of SRC-3 inhibited cell proliferation, affected cell cycle progression (Supplementary information, Figures S1 and S2) and abolished the induction of PRs (PR-A and PR-B) by estradiol, an endogenous target gene of ER, confirming the functional importance of SRC-3 (Figure 1B, compare lanes 2 and 4). After the successful knockdown of SRC-3, the cells were immediately transduced with a lentiviral cDNA expression library, followed by antibiotic selection and treatment with doxorubicin to further reduce cell survival (Figure 1C). A total of 34 surviving and proliferating clones were isolated for recovery of cDNAs. Sequencing analyses revealed that some cDNAs encode the same genes, and a total of 10 genes were identified. Among the 10 genes identified, 8 genes belong to the ribosomal biogenesis (5 genes) and the cytokine/mitogen (3 genes) signaling pathways, and have been implicated in cancer development (data not shown) 37. In addition, cathepsin D, another well-known target gene of ER whose overexpression has been associated with increased risk of relapse and metastasis was also isolated. These results indicate that our screening strategy selectively identified genes that are important for cell survival and may be involved in the oncogenic function of SRC-3.
SRC-3 regulates MIF expression. (A) Immunoblot analysis demonstrated specific knockdown of SRC-3, but not SRC-1 or SRC-2, in MCF-7 cells by infection with lentivirus expressing shRNA against SRC-3. (B) Knockdown of SRC-3 abolished the induction of PRs (PR-A and PR-B) by estradiol. (C) Schematic representation of the strategy used to identify genes that promoted cell survival in MCF-7 breast cancer cells under SRC-3 knockdown condition. (D) Immunoblot analysis showed that compared to transfection with control siRNA, transfection of siRNAs against SRC-3 (siSRC-3) suppressed MIF protein expression. β-actin was used as loading controls. (E) Semi-quantitative RT-PCR showed that the MIF mRNAs were reduced in SRC-3 knockdown cells. GAPDH was used as loading controls. (F) Expression of MIF protein in mammary epithelial cells derived from SRC-3-null mutant (SRC-3–/–) mice was reduced compared to wild-type cells. β-actin was used as loading control. (G) Levels of Mif mRNA from MEFs and primary lymphocytes from wild-type (SRC-3+/+) or null mutant (SRC-3–/–) mice were compared by RNase protection assay (RPA). L32 and GAPDH were used as loading controls. All normalized intensity is quantified by ImageJ.
In the current study, we focused on one of the candidate genes MIF (accession number NM_002415), a cytokine and a growth factor, to investigate how regulation of MIF expression by SRC-3 may contribute to cancer development. Similar to SRC-3, MIF was also overexpressed in patients with cancer 14,15,16,17. Despite being overexpressed in cancer patients, the molecular mechanism responsible for MIF overexpression is not well understood. Given the fact that MIF was identified from the screen, we hypothesize that SRC-3 regulates MIF expression and MIF promotes cell survival.
We first examined whether expression of MIF is regulated by SRC-3. In fact, knockdown of SRC-3 in MCF-7 cells reduced MIF expression at the protein and mRNA levels as detected by immunoblot or RT-PCR analysis, respectively (Figure 1D and 1E). A similar decrease in endogenous MIF protein and Mif mRNA expression was found in cells derived from SRC-3-null mutant mice, including decreased protein expression in the mammary epithelial cells (Figure 1F) and reduced abundance of mRNA transcripts in the primary MEFs and lymphocytes (Figure 1G). These results indicated that regulation of MIF expression by SRC-3 occurs at the transcriptional levels and this regulation is not limited to the MCF-7 breast cancer cells.
Activation of the MIF promoter by SRC-3 requires phosphorylation of SRC-3 and the hypoxia responsive element on the MIF promoter
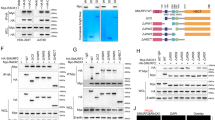
To further understand how SRC-3 regulated MIF expression at the transcriptional level, we cloned a 5′ flanking region (from −2 504 to +127 bp) of the human MIF gene and used it to perform luciferase reporter gene assays. To further map the region of the MIF promoter responsive to SRC-3, a series of 5′ truncated luciferase reporter constructs, including those starting at −1 514, −614, −434, −254 and −74 were generated (Figure 2A). Results from the reporter gene assays showed that co-transfection of SRC-3 activated the full-length as well as the five promoter truncation variants (Figure 2A). These results identified the promoter region from −74 to +127 bp is required for activation by SRC-3 (Figure 2A).
HRE on the MIF promoter and phosphorylation of SRC-3 are required for the activation by SRC-3. (A) MCF-7 cells were transfected with MIF promoter-regulated luciferase reporter combined with either control vector or with SRC-3 expression vector. The luciferase activity was measured 48 h after transfection and normalized against total input protein. The normalized activity from vector-transfected sample was set as 1. The values represented fold activation over the control and were expressed as means ± SD from three independent experiments performed in duplicate. (B) Wild-type MIF promoter or promoter with mutation at HRE or CRE was co-transfected with expression vector for wild-type SRC-3 into the MCF-7 cells. The luciferase activity was determined as in A. Shown are means ± SD from three experiments performed in duplicate. (C) MCF-7 cells were transfected with the MIF promoter, together with the wild-type SRC-3 or the indicated SRC-3 mutant. The luciferase activity was determined as described in A. A schematic of the phosphorylation sites on SRC-3 and a representative immunoblot analysis were shown. The values are means ± SD from three experiments performed in duplicate.
Within this promoter region (−74 to +127 bp), a putative hypoxia responsive element (HRE) and a cyclic adenosine monophosphate response element (CRE) were predicted by PROMO (http://alggen.lsi.upc.es/recerca/menu_recerca.html) and JASPAR (http://jaspar.cgb.ki.se). Having established that SRC-3 activated the MIF promoter, we sought to determine which element is required. Our results showed that when compared to the wild-type MIF promoter, mutation of the HRE, but not the CRE, significantly reduced the activation by SRC-3 (Figure 2B). These results indicated that activation by SRC-3 required the HRE on the MIF promoter.
Since phosphorylation was previously shown to be important for the coactivator function and specificity of SRC-3 8, 38, 39, we investigated the role of SRC-3 phosphorylation in the activation of the MIF promoter. For this purpose, we examined the ability of six SRC-3 phosphorylation mutants to activate the MIF promoter. Each mutant contains single amino acid mutation to alanine at previously identified phosphorylation sites and are herein referred to as T24A, S505A, S543A, S857A, S860A and S867A (Figure 2C, top left panel) 8. We found that although most mutations did not affect the activity of SRC-3 on the MIF promoter, mutation of T24 (T24A) resulted in an ∼40% loss of activation; and mutation of S857 (S857A) abolished the activation by SRC-3 (Figure 2C, right panel). Since all the mutants were expressed at levels similar to wild-type SRC-3 (Figure 2C, bottom left panel), our results suggested that phosphorylation of S857 regulates the ability of SRC-3 to activate the MIF promoter. Interestingly, we found that only SRC-3, but not SRC-1 or SRC-2, was able to activate the native MIF promoter, underscoring coactivator specificity (Supplementary information, Figure S3).
Phosphorylation of S857 on SRC-3 by IKKα is important for the activation of the MIF promoter
The activity of SRC-3 is known to be regulated by phosphorylation 8, 38, 39. Specifically, our results showed that phosphorylation of S857 on SRC-3 is required for the activation of MIF promoter (Figure 2C). Since S857 of SRC-3 was previously shown to be phosphorylated by IKKs 8, 40, 41, we investigated the involvement of IKKα and IKKβ in activation of the MIF promoter. By co-transfection with the MIF promoter, we found that the IKKα catalytic subunit activated the MIF promoter better than the IKKβ subunit (Figure 3A). In addition, IKKα synergistically activated the MIF promoter when co-transfected with the wild-type SRC-3, but not the S857A mutant (Figure 3B, columns 5 and 6). Similar to knockdown of SRC-3, knockdown of IKKα by siRNA reduced MIF expression (Figure 3C). Our current data are consistent with our earlier report, which showed that IKKα but not IKKβ phosphorylated and activated SRC-3 40, and suggested that phosphorylation of S857 on SRC-3 by IKKα is important for the activation of the MIF promoter.
Phosphorylation of S857 on SRC-3 by IKKα is important for the activation of the MIF promoter. (A) MCF-7 cells were transfected with the MIF promoter together with vector control, or expression vector for IKKα or IKKβ. The luciferase activity was determined as previously described. Expression of IKKα and IKKβ was shown. A non-specific band was indicated by n.s. The values are mean ± SD from three independent experiments performed in duplicate. (B) MCF-7 cells were transfected with the MIF promoter together with control vector or the indicated expression vectors. The luciferase activity was determined as previously described. The values are mean ± SD from three independent experiments performed in duplicate. (C) Reduced levels of MIF after knockdown of IKKα or SRC-3 were shown. Knockdown of SRC-3, IKKα or MIF was confirmed. β-actin was used as loading controls.
Phosphorylation of S857 is required for the coactivation of the MIF promoter by SRC-3, CBP and hypoxia-inducible factor-1α
Recently, genome-wide mapping has identified potential high-stringency hypoxia-inducible factor (HIF)-binding site(s) near the MIF gene 42. Accordingly, the HIF-1alpha (HIF-1α) transcriptional factor has been reported to induce MIF expression by binding to the HRE of the MIF promoter 18, 43. Similarly, we found that activation of the MIF promoter region (−74 to +127 bp) by HIF-1α is dependent on the presence of a functional HRE but not the CRE, and HIF-1α was recruited to this region of the MIF promoter (Supplementary information, Figure S4). As a transcriptional coactivator, SRC-3 is recruited to promoters through interactions with transcriptional factors. Since activation of the MIF promoter (−74 to +127 bp) by SRC-3 also required a functional HRE (Figure 2B), these results suggest that SRC-3 collaborates with HIF-1α to regulate MIF expression. Therefore, we determined whether SRC-3 could function as a coactivator for HIF-1α. In fact, co-transfection of HIF-1α with wild-type SRC-3 resulted in a synergistic activation of the MIF promoter compared to transfection of either one alone (Figure 4A, columns 2, 4 and 7). Similar synergistic activation can also be observed in HeLa and PC3 cells, and is not limited to MCF-7 breast cancer cells (Supplementary information, Figure S5). On the other hand, transfection of an S857A mutant, either alone or in combination with HIF-1α, failed to activate the same MIF promoter (Figure 4A, columns 5 and 8).
Phosphorylation of S857 is important for the activation of human MIF promoter by SRC-3, CBP and HIF-1α. (A) MCF-7 cells were transfected with the MIF promoter, expression vectors for HIF-1α, wild-type SRC-3 or S857A mutant, either alone or combined as indicated. The luciferase activity was determined as previously described. Shown is the mean ± SD from three experiments performed in duplicate. (B) Cell lysates from MCF-7 cells were used to determine the interaction between endogenous SRC-3 and HIF-1α by co-immunoprecipitation (Co-IP; left panel). HEK293T cells transfected with HIF-1α, alone or together with wild-type SRC-3 or S857A mutant as indicated, were used for Co-IP experiments. Top right panel showed the levels of HA-HIF-1α co-immunoprecipitated with Flag-SRC-3 (middle panel). Bottom panel showed equal expression of HIF-1α. Shown is a representative from three experiments with similar results. Normalized intensity is quantified by ImageJ. (C) MCF-7 cells were transfected with the MIF promoter, expression vectors for HIF-1α, CBP, wild-type SRC-3 or S857A mutant, either alone or combined as indicated. The luciferase activity was determined as previously described. Shown is the mean ± SD from three experiments performed in duplicate. Due to higher number (four) and more total amount of plasmids transfected, less activation by SRC-3 was observed under this particular condition. (D) The MCF-7 cells were first transfected with siRNAs to knockdown the protein indicated. The next day, these cells were transfected as described in C. The luciferase activity was also determined as described in C. (E) The lysates from MCF-7 cells transfected with the indicated siRNAs either alone or in different combination were used for immunoblot to determine the levels of endogenous MIF. β-actin was used as loading controls.
To further understand how phosphorylation of S857 on SRC-3 affects its ability to regulate MIF expression, we next determined whether SRC-3 interacts with HIF-1α, and whether phosphorylation of S857 affected interaction between SRC-3 and HIF-1α. Our co-immunoprecipitation results showed that indeed both endogenous SRC-3 and transfected wild-type SRC-3 interacted with HIF-1α (Figure 4B). However, mutation of S857 reduced but did not completely abolish the interaction of SRC-3 with HIF-1α (Figure 4B), suggesting that phosphorylation of S857 'enhanced' the interaction of SRC-3 with HIF-1α. We were aware that this partially reduced interaction could not fully explain the complete loss of activation by the S857A mutant and suggested involvement of other coactivators.
CBP is a part of SRC-3 coactivator complex and contains the HAT activity essential for transcription activation. Previously, we showed that phosphorylation of S857 on SRC-3 is required for its interaction with CBP 8. Since our current results also showed that phosphorylation of S857 on SRC-3 is required for the activation of MIF promoter (Figure 2C), we investigated the involvement of CBP in this activation. Our results showed that when co-expressed, CBP was able to collaborate with HIF-1α and wild-type SRC-3, and further activated the MIF promoter (Figure 4C, columns 2, 7 and 8). In contrast, no collaboration of CBP with the S857A mutant was observed (Figure 4C, columns 4 and 9).
To further demonstrate that HIF-1α, SRC-3 and CBP are all required for the expression of MIF, we determined the effects of siRNA knockdown of either one or two, or all three of these proteins on the activation of MIF promoter and MIF protein expression. Our results showed that knockdown of endogenous SRC-3 or CBP, either alone or SRC-3 and CBP combined, inhibited the activation of the MIF promoter even in the presence of HIF-1α overexpression (Figure 4D, compare lanes 2, 4, 6 and 8). Similarly, knockdown of endogenous HIF-1α or CBP, either alone or HIF-1α and CBP combined, inhibited the activation of MIF promoter by overexpression of SRC-3 (Figure 4D, compare lanes 10, 12, 14 and 16). In line with these results, our ChIP results showed that by overexpression, recruitment of HIF-1α, SRC-3 and CBP to the region on the endogenous MIF promoter that contains the HRE and is responsible for the activation by HIF-1α and SRC-3 was enhanced (Supplementary information, Figure S6). Finally, our results further showed that knockdown of HIF-1α, SRC-3 or CBP alone reduced the endogenous levels of MIF (Figure 4E, lanes 1, 2, 3 and 8), and knockdown of two or three proteins combined further reduced the MIF protein levels (Figure 4E, lanes 4, 5, 6 and 7, and Supplementary information, Figure S7).
These results identified both SRC-3 and CBP as potential coactivators for HIF-1α, and indicated that phosphorylation of S857 on SRC-3 is required for the recruitment of CBP and is essential for the coactivation of the MIF promoter by SRC-3 and CBP. Taken together, we demonstrated that loss of SRC-3 function (by knockdown and knockout of SRC-3) reduced endogenous MIF expression at mRNA and protein levels in several different cell types (Figure 1). We also demonstrated that gain of SRC-3 function activates the MIF promoter in the promoter assays (Figure 2A). In addition, we identified that phosphorylation of a specific amino acid residue (S857) on SRC-3 by IKKα is required for the activation of MIF promoter (Figures 2C and 3). Finally, we demonstrated that co-activation of the MIF promoter by transcription factor HIF-1α, SRC-3 and CBP (Figure 4). Our data suggest that SRC-3 regulates MIF expression by functioning as a coactivator of HIF-1α.
MIF regulates autophagy
The ability of SRC-3 to promote cancer cell survival is clinically important, but the mechanism remains poorly understood. Having established that SRC-3 regulated MIF expression, we investigated the potential role of MIF in cell survival. In addition to apoptosis, autophagic cell death is another major mechanism that plays a tumor suppressor function 26,27,28. Since knockdown of SRC-3 or MIF reduced Bcl-2 levels, but did not significantly affect apoptosis (Supplementary information, Figure S8), we investigated whether MIF regulated autophagy. Cleavage of the autophagy marker microtubule-associated protein 1 light chain (LC3) to LC3-I and conjugation of LC3-I to phosphatidylethanolamine to form LC3-II is an essential process for the formation of autophagosomal vacuoles 44,45,46,47,48,49. As the levels of LC3-II are proportional to the extent of autophagic vacuole formation, we therefore examined the processing of LC3 as an indicator of autophagy activity. To this end, we generated a line of MCF-7 cells that stably expressed the autophagy marker LC3 fused with GFP (GFP-LC3). Expression of the GFP-LC3 fusion protein is a widely used autophagy assay, which allows visualization of autophagosome formation in live cells, because GFP-LC3 is processed and recruited to the autophagosome membrane where it can be imaged as a cytoplasmic punctate structure by fluorescence microscopy 46, 50, 51.
In contrast to control siRNA-treated cells that exhibited a diffuse cytoplasmic GFP-LC3 pattern, knockdown of SRC-3 or MIF resulted in a change of intracellular localization of GFP-LC3 and formation of punctate structures, indicative of autophagic activity (Figure 5A). The punctate structures from SRC-3 or MIF knockdown cells are very similar to that found in cells treated with rapamycin, a known inducer of autophagy (Supplementary information, Figure S9). Quantification showed that the percentage of GFP-LC3 punctate-positive cells increased from 3% in control siRNA-transfected cells to 32% and 25% in cells treated with siRNAs against SRC-3 and MIF, respectively (Figure 5B). To further demonstrate that knockdown of SRC-3 or MIF promotes autophagy, we determined the expression of endogenous autophagy-specific LC3-II. In agreement with imaging of GFP-LC3 in live cells, quantification of the immunoblot analysis revealed an elevated level of autophagy-specific LC3-II expression in cells that were treated with specific siRNAs against SRC-3 or MIF, compared to cells treated with control siRNA (Figure 5C). In addition, the endogenous levels of sequestosome 1 (SQSTM1/p62) were significantly reduced in the SRC-3 or MIF knockdown cells (Figure 5C). Since SQSTM1/p62 is degraded by autophagy 52,53,54, these results support that knockdown of SRC-3 or MIF elevated autophagy activity.
SRC-3 and MIF regulate autophagy. (A) Induction of autophagy in SRC-3 or MIF knockdown MCF-7 cells was visualized by the formation of punctated GFP-LC3 structure. MCF-7 cells that stably expressed the GFP-LC3 fusion were transfected with either control siRNA or siRNAs specific for SRC-3 or MIF. The cells were imaged 36 h after transfection. (B) Quantification of autophagy represented the ratio of GFP-LC3 punctate-positive cells to the total cells counted. More than 300 cells per group were counted in each experiment and the results were from three independent experiments. (C) Cell lysates from MCF-7 transfected with control siRNA or siRNAs specific against SRC-3 (siSRC-3) or MIF (siMIF) were analyzed with antibodies against LC3 or SQSTM1/p62 to determine the autophagy activity. Knockdown of SRC-3 or MIF was confirmed with specific antibodies. β-actin was used as loading controls. Normalized intensity is quantified by ImageJ. Graph shows quantification data in arbitrary units for the density of the LC3-II (top right) or SQSTM1/p62 (bottom right) bands from each sample. Columns and bars represent the mean ± SD of the results from three independent experiments. Difference is considered significant at the 95% confidence level.
Despite an earlier report indicating that MCF-7 cells did not express beclin 1, the essential mediator for autophagy, and were deficient in autophagy 55, numerous reports have since described beclin 1 expression in MCF-7 cells and showed that MCF-7 cells were indeed autophagy competent 56,57,58,59. In fact, we found that beclin 1 was expressed in our MCF-7 cells at levels comparable with other cells and that the cells were autophagy-competent (Supplementary information, Figure S10). The confirmation of beclin 1 expression allowed us to study regulation of autophagy by SRC-3 or MIF in MCF-7 cells. It is of note that induction of autophagy is not limited to MCF-7 cells, as increased punctate GFP-LC3 formation and elevated LC3-II levels were similarly observed in HeLa and MDA-MB-231 cells after knockdown of SRC-3 or MIF (data not shown).
MIF regulates cell viability through autophagy
Having demonstrated that SRC-3 and MIF regulated autophagy, we hypothesized that MIF may mediate important oncogenic functions of SRC-3 through its regulation of autophagy. Consistent with this hypothesis, our results showed that knockdown of MIF phenocopied knockdown of SRC-3 and inhibited cell proliferation (Figure 6A and Supplementary information, Figures S1 and S2), and reduced colony number in a colony-formation assay (Figure 6B and 6D, top row). Interestingly, transfection of SRC-3 that is resistant to the shRNA targeting the SRC-3 3′-UTR into the SRC-3 knockdown cells (sh3′-UTR, Figure 6C, lanes 3 and 4) increased the colony number (Figure 6B) and elevated the expression levels of endogenous MIF (Figure 6C, lanes 3 and 4). Since colony-formation assay is based on the ability of a single cell to grow into a colony, these results indicated that knockdown of SRC-3 or MIF reduced cell viability and MIF may, at least partially, mediate this important function of SRC-3.
MIF regulates cell viability through autophagy. (A) Proliferation of MCF-7 cells after knockdown of SRC-3 (shSRC-3) or MIF (shMIF) was compared to the control cells (shControl). The cell numbers at 2, 4 and 6 days after plating were measured by MTT assay. The values represent means ± SD from three experiments. (B) The viability of the cells transfected with control vector or SRC-3 expression construct after knockdown of SRC-3 by shRNA targeting the 3′-UTR of SRC-3 was determined by colony-formation assay. (C) The expression of transfected Flag-SRC-3 and endogenous MIF from experiments described in B was determined by immunoblot analysis in a parallel experiment. β-actin was used as loading controls. (D) The viability of MCF-7 cells after knockdown of SRC-3 or MIF was determined by colony-formation assays. The medium was changed every 2 days. For treatment with MIF, MIF (50 ng/ml) was added to the medium every time the medium was changed. The colonies were then fixed and stained with 0.5% crystal violet. Shown is a representative from three experiments with similar results. (E) MCF-7 cells were transfected with control (siControl) or siRNA specific for SRC-3 (siSRC-3). After 48 h of transfection, the cells were treated with MIF (50 ng/ml) for an additional 4 h when indicated. Cell lysates were analyzed using the indicated antibody. β-actin was used as loading controls. (F) MCF-7 cells were transfected with control (siControl) or siRNA specific for MIF (siMIF), and treated as in C. Cell lysates were analyzed by immunoblot using the indicated antibody. β-actin was used as loading controls. All normalized intensity is quantified by ImageJ. Graph shows quantification data in arbitrary units for the density of the LC3-II or SQSTM1/p62 bands from each sample. Columns and bars represent the mean ± SD of the results from three independent experiments. Difference is considered significant at the 95% confidence level.
Consistent with this hypothesis, we found that addition of biologically active recombinant human MIF was able to significantly rescue cell death and increased colony number following knockdown of SRC-3 or MIF (Supplementary information, Figure S11 and Figure 6D, bottom row). In line with its ability to rescue cell death, treatment with MIF also inhibited the induction of endogenous autophagy-specific LC3-II and elevated the SQSTM1/p62 levels in the SRC-3 or MIF knockdown cells (Figure 6E and 6F). Our results substantiated that knockdown of SRC-3 or MIF reduced cell viability through induction of autophagic cell death and demonstrated that treatment of MIF inhibited autophagic cell death induced by knockdown of SRC-3 or MIF. The fact that MIF is able to inhibit autophagy and promote cell survival, not only in MIF knockdown but also in SRC-3 knockdown cells, underlines the importance of MIF regulation by SRC-3.
Downregulation of MIF suppressed tumorigenesis and enhanced chemosensitivity
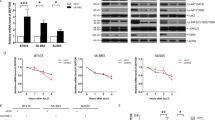
To evaluate the functional importance of autophagic cell death induced by MIF knockdown, we determined its effects on the tumorigenicity of MCF-7 cells in a tumor xenograft mouse model 60,61,62. In order to maintain long-term knockdown of MIF after injection into mice, we used the MCF-7 cells with stable knockdown of MIF. Consistent with results from transient MIF knockdown (Figure 6F), stable knockdown of MIF in MCF-7 cells also increased the expression of autophagy-specific LC3-II and decreased the levels of SQSTM1/p62 (Figure 7A, a). Conversely, treatment with MIF reduced the induction of LC3-II expression and increased the levels of SQSTM1/p62 (Figure 7A, a). We then injected athymic nude mice (five mice per group) with either the control (shControl) or the stable MIF knockdown (shMIF) cells. After injection, the mice were monitored for development of tumors and the tumor sizes were measured. Remarkably, all five mice that were injected with the shControl cells developed tumors 3 weeks after injection and the sizes of the tumors increased 5 weeks after injection (Figure 7A, b). In contrast, all five mice that were injected with the shMIF cells failed to develop visible tumors during the same period (Figure 7A, b and c). A representative image comparing the mice 5 weeks after injection clearly demonstrated the difference in tumor development (Figure 7A, c). All mice were sacrificed 5 weeks after injection due to the large size of tumors in the control mice (Figure 7A, d). We noticed that all five mice that contained tumors also developed enlarged spleens (Figure 7A, e). These results clearly demonstrated that knockdown of MIF inhibited the tumorigenicity of MCF-7 cells in a tumor xenograft mouse model.
Downregulation of MIF suppressed tumorigenicity and enhanced chemosensitivity in MCF-7 breast cancer cells. (A) a. Expression of autophagy-specific LC3-II and SQSTM1/p62 in stable MIF knockdown cells was determined. When indicated, cells were treated with MIF (50 ng/ml) for 4 h before harvest. Normalized intensity is quantified by ImageJ. b. Comparison of tumor volumes from mice injected with shControl or shMIF cells measured at 3 and 5 weeks after injection. Gray and black bars represent tumor sizes from the two injected sides. c. A representative image of the mice at week 5 post-injection. d. Tumors taken from the siControl cells-injected mice (panel d). e. A representative image showed the spleen from the siControl cells injected mice was enlarged. (B) Stable MIF knockdown MCF-7 cells were treated with 0.1, 1 and 10 μM of doxorubicin or etoposide for 18 h, and the cell viability was determined by MTT assay. Shown is mean ± SD from three experiments performed in triplicate. (C) Stable MIF knockdown MCF-7 cells were treated with low concentrations (0.1 and 1 μM) of doxorubicin or etoposide. The levels of autophagy-specific LC3-II were determined. Knockdown of MIF was confirmed. β-actin was used as loading controls. Normalized intensity is quantified by ImageJ. Graph shows quantification data in arbitrary units for the density of the LC3-II or SQSTM1/p62 bands from each sample. Columns and bars represent the mean ± SD of the results from three independent experiments. Difference is considered significant at the 95% confidence level.
Since chemotherapy is used to treat all stages of breast cancer, and resistance of cancer cells to chemotherapy is the major cause for subsequent cancer recurrence and metastasis, we hypothesized that autophagic cell death induced by MIF knockdown may enhance chemosensitivity of cancer cells. To test this hypothesis, we compared the cytotoxicity of doxorubicin and etoposide after knockdown of MIF. Our results showed that doxorubicin and etoposide reduced the viability of control cells in a dosage-dependent manner (Figure 7B, shControl). Importantly, knockdown of MIF enhanced the cytotoxicity of doxorubicin and etoposide as evidenced by the further decrease in cell viability compared to the control cells (Figure 7B, shMIF). In contrast, overexpression of MIF reduced the chemosensitivity to doxorubicin and etoposide and promoted cell survival and proliferation (Supplementary information, Figure S12). To confirm that knockdown of MIF sensitized cells to chemotherapeutic killing by induction of autophagy, we examined the levels of autophagy-specific LC3-II after treatment with low concentrations of doxorubicin or etoposide. Immunoblot analysis showed that the levels of LC3-II were not affected in the control cells treated with low concentrations (0.1 and 1 μM) of doxorubicin or etoposide (Figure 7C, shControl, lanes 1-5). However, knockdown of MIF increased the levels of LC3-II as expected (Figure 7C, shMIF, lane 6), and treatment with low concentrations of doxorubicin or etoposide further increased the levels of LC3-II (Figure 7C, compare lane 6 with lanes 7-10). Once again, we found that apoptosis was not significantly affected in the MIF knockdown cells compared to control cells (Supplementary information, Figure S13). Together, our results indicated that by regulating autophagic cell death, MIF plays an important role in both tumorigenesis and chemosensitivity of cancer cells.
Discussion
SRC-3 is an oncogene that is involved in the initiation and progression of many cancers, including breast and prostate cancers 63, 64. Recently, clinical evidence suggested that SRC-3 promoted drug resistance in cancer cells and contributed to poor disease-free survival in patients with breast and lung cancers 3,4,5. Therefore, a better understanding of how SRC-3 promotes drug resistance and survival in cancer cells is imperative for improving treatment outcomes.
In this report, we utilized a chemotherapeutic sensitivity and cDNA rescue assay to identify MIF as a target gene of SRC-3, thereby uncovering a novel function of SRC-3 and MIF in cell death. MIF, a cytokine and a growth factor, has multiple biological functions and is known to play an important role in inflammation and cancer 9,10,11, 13, 65. We found that expression of MIF is regulated by SRC-3 at the transcriptional level as demonstrated by reduced Mif mRNA in cells derived from SRC-3-null mutant mice, including MEFs and primary lymphocytes, and transfection of SRC-3 activated the MIF promoter in MCF-7 cells. We showed that the region from −74 to +127 bp of the MIF promoter is required for activation by SRC-3. This region contains a putative HRE and a CRE. Mutation of the HRE, but not the CRE, within the promoter significantly reduced the activation by SRC-3, suggesting that SRC-3 cooperates with HIF-1α to activate MIF expression. In fact, we showed that SRC-3 and HIF-1α synergistically activated the MIF promoter in different cell types, and further identified phosphorylation of S857 on SRC-3 by IKKα is required for this synergistic activation. Although phosphorylation of S857 is required for the activation of MIF promoter by SRC-3, it is not required for the interaction of SRC-3 with HIF-1α, suggesting that phosphorylation of S857 by IKKα recruits other important coregulators to activate the MIF promoter. To this end, we showed that CBP collaborated with SRC-3 and HIF-1α to activate the MIF promoter, and that co-activation by CBP is dependent on the phosphorylation of S857on SRC-3. These results are in line with a previous report, which showed that phosphorylation of S857 was required for the direct interaction of SRC-3 and CBP 8. Based on these results and the fact that HIF-1α, SRC-3 and CBP can be recruited to the endogenous MIF promoter, transcriptional activation of MIF requires not only the binding of HIF-1α to the MIF promoter, but also the recruitment of crucial coactivators SRC-3 and CBP (Figure 8). SRC-3 is recruited through direct interaction with HIF-1α that can occur independent of the phosphorylation status of S857 on SRC-3, although phosphorylation of S857 appears to enhance this interaction. Thus, phosphorylation of S857 provides a platform for SRC-3 to further recruit CBP that is essential for the activation of the MIF promoter.
A model for activation of MIF by the HIF-1/SRC-3/CBP complex. Activation of MIF expression requires the binding of HRE by HIF-1α, which in turn recruits SRC-3 to the MIF promoter. Phosphorylation of S857 on SRC-3 by IKKα is required for the subsequent recruitment of CBP and eventual activation of MIF promoter. The ability of MIF to regulate autophagic cell death underscores its potential importance in cancer development.
It was previously shown that expression of MIF is induced by HIF-1α under hypoxia, and that induction of MIF prevents premature senescence by inactivating the p53 tumor suppressor 18. Although recent genome-wide analysis of SRC-3 chromatin affinity sites in MCF-7 human breast cancer in response to estrogen induction did not identify the MIF gene, high-stringency HIF-binding site(s) near the MIF gene was identified in another recent genome-wide mapping 42, 66. It is reasonable that as a master transcriptional regulator, the ability of SRC-3 to regulate expression of its target genes is dependent on the transcriptional factors involved (i.e., HIF-1 vs ERα). Since intratumoral hypoxia is a major obstacle to cell survival and MIF expression is regulated by oncogene SRC-3, we hypothesized that MIF may be an important mediator of cell survival and the oncogenic function of SRC-3. In support of this hypothesis, we found that knockdown of MIF phenocopied knockdown of SRC-3 and resulted in not only inhibition of cell proliferation, but also reduction of cell viability. Importantly, our data further showed that knockdown of MIF induced autophagy, inhibited tumorigenicity and enhanced the cytotoxicity of doxorubicin and etoposide in MCF-7 breast cancer cells. However, it should be pointed out that both intracellular and extracellular MIF are functionally important and it is difficult to distinguish the functions of these two pools of MIF, because secreted MIF could work in autocrine and paracrine fashion.
In contrast to autophagy, apoptotic cell death was not significantly affected by knockdown of SRC-3 or MIF, despite the fact that expression of the anti-apoptotic Bcl-2 protein was inhibited. This is consistent with recent reports that treatment of MCF-7 cells with Bcl-2 siRNA significantly reduced cell viability (50%-85%), but only slightly induced apoptosis (9%-11%), indicating that another cell death mechanism may be involved 56, 67, 68. Interestingly, Bcl-2 also inhibited the beclin-1-dependent autophagy by binding to beclin 1. Consequently, it is likely that knockdown of SRC-3 or MIF suppressed Bcl-2 expression, which in turn, removed its inhibition of beclin 1 and activated autophagy. Future study to understand the detailed mechanism by which SRC-3 and MIF regulate autophagy is warranted and ongoing.
It should be noted that although SRC-1 and SRC-2 have been shown to interact with HIF-1α and activate a synthetic promoter with tandem HREs under hypoxia 69, we found that SRC-3, but not SRC-1 or SRC-2, activated the native MIF promoter. Consistent with our findings, a recent report showed that knockdown of SRC-3, but not SRC-1 or SRC-2, reduced erythropoietin and VEGF induction in Hep3B cells by HIF-2α and HIF-1α, respectively 70. Since SRC-3, but not SRC-1 or SRC-2, has been shown to be phosphorylated by IKKα at S857 and is required for the activation of the MIF promoter, it is possible that selective phosphorylation regulates the functional specificity of coactivator complex assembly 40, 41.
Although the circumstances for this coactivator specificity remain unknown, these results substantiated that the function of the p160/SRC family of coactivators is not redundant and that different HIF target genes exhibit different requirements for specific members of the p160/SRC. Most importantly, we have demonstrated that expression of MIF is regulated by the oncogene SRC-3, and identified 'suppression of autophagy' as a novel mechanism by which SRC-3 and MIF increase the chemoresistance and tumorigenicity (Figure 8). By virtue of its ability to regulate MIF expression, the role of SRC-3 in autophagic cell death is mediated through regulation of MIF expression. Our study provides the basis for targeting the SRC-3/MIF-autophagy pathway to improve the therapeutic outcome for breast cancer patients.
Materials and Methods
Cell lines and culture conditions
MCF-7, MDA-MB-231, HeLa, PC3 and HEK293T cells were obtained from the American Type Culture Collection and were maintained as instructed. The MEFs, primary lymphocytes and the mammary epithelial cells have been described previously 8, 71.
Plasmids and reagents
The mammalian expression vectors for SRC-3, CBP and IKKs plasmids have been described previously 8, 41. The human HIF-1α cDNA was purchased from Open Biosystems and subcloned into a pSG5-HA or pSG5-Flag vector modified from pSG5 vector (Stratagene). The MIF cDNA was inserted into the pCMV-Tag 2B vector (Stratagene). The full-length human MIF promoter (−2 504 to +127 bp) was amplified from genomic DNA prepared from MCF-7 cells and cloned into the pGL3-basic vector (Promega). Truncated MIF promoter variants were constructed by PCR amplification using the full-length human MIF promoter as a template. The following primers were used. MIF promoter −2 504 sense 5′-CCGCTCGAGCTTTCCCAAGGAACTAAGGCTGAGCCAAG-3′. MIF promoter −1 514 sense 5′-CCGCTCGAGTCAGTCTTCTGCCTGGAGTGACTT-3′. MIF promoter −614 sense 5′-CCGCTCGAGGGAGCTGAGCACGTTTGAACCACT-3′. MIF promoter −434 sense 5′-CCGCTCGAGACAAGCTCACGCATGCGGGACTGG-3′. MIF promoter −254 sense 5′-CCGCTCGAGCGGTGACTTAGTGAAAGGACTAAG-3′. MIF promoter −74 sense 5′-CCGCTCGAGTCCCCACTCGGGGCGGAGCCGCAG-3′. The same anti-sense primer was used 5′-CCCAAGCTTGGCATGATGGCAGAAGGACCAGGAGACCC-3′.
The GFP-LC3 was generated by inserting the LC3 cDNA obtained from MCF-7 by RT-PCR into the pEGFP-C3 vector (Clontech).
Doxorubicin, etoposide and (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester (ISO-1) were obtained from Calbiochem. M2 beads and β-actin antibodies were from Sigma. LC3 antibody was from MBL. MIF, p21 and IKKα antibodies were from Santa Cruz Biotechnology. HA antibody was from Roche. Rabbit polyclonal antibodies against SRC-3 have been described previously 41. CBP and HIF-1α antibodies were from Upstate and BD Biosciences, respectively. SQSTM1/p62, antibodies against phosphorylated ERK and total ERK were from Cell Signaling Technology. Recombinant human MIF were from R&D systems and eBioscience. Puromycin and blasticidin were purchased from InvivoGen. Rapamycin was from Selleck Chemicals. All oligonucleotides were from Invitrogen.
Assay to identify genes that can promote cell survival in SRC-3 knockdown cells
Knockdown of SRC-3 was achieved by infecting the MCF-7 breast cancer with lentivirus expressing shRNA against SRC-3. After successful knockdown of SRC-3, the cells were transduced with a lentiviral cDNA expression library, followed by selection with blasticidin (10 μg/ml) and treatment with doxorubicin (10 μg/ml) until surviving colonies were clearly visible. A total of 34 survival clones were isolated for recovery of cDNAs.
Sequencing analyses were performed to identify the candidate genes.
Knockdown of SRC-3 by lentivirus expressing shRNA against SRC-3 or MIF
Knockdown of SRC-3 or MIF was achieved by infecting the MCF-7 breast cancer cells with lentivirus expressing shRNA against SRC-3 or MIF. To produce shSRC-3- or MIF-expressing lentivirus, the plasmids were co-transfected with pPACKH1-packaging mix into 293TN producer cells according to manufacturer's instructions (SBI). Collection of the produced virus and transduction of the MCF-7 breast cancer cells with the virus were also performed according to manufacturer's instructions (SBI). For stable selection, puromycin (8 μg/ml) was added to the medium. The medium was changed every 3 days until colony was clearly visible. To generate shRNA for stable knockdown, the specific sequences against SRC-3 (5′-TCGAGACGGAAAACATTGTA-3′), SRC-3 3′-UTR (5′-AACACTGCACTAGGATTATTG-3′) and MIF (5′-TCATCGTAAACACCAACGT-3′ and 5′-GCGCAGAACCGCTCCTACA-3′) were inserted into the pSIH-H1-Puro vector (SBI) according to manufacturer's instructions.
Lentiviral cDNA library
The cDNA library was constructed using the CloneMiner II cDNA Library Construction Kit (Invitrogen) according to manufacturer's instructions. The finished library contains 1.2 × 106 clones and the average cDNA size is 1.3 kb.
To produce lentiviral cDNA library, the plasmids were co-transfected with pPACKH1-packaging mix into 293TN producer cells according to manufacturer's instructions (SBI). Collection of the produced virus and transduction of target cells with the virus were also performed according to manufacturer's instructions (SBI). For stable selection, blasticidin (10 μg/ml) was added to the medium. The medium was changed every 3 days until colony was clearly visible.
Knockdown of gene by transfection with siRNAs against SRC-3, MIF, or IKKα
All the ON-TARGETplus siRNA reagents were purchased from Dharmacon. Transfection of siRNAs by TransIT-TKO transfection reagent was performed according to manufacturer's instructions (Mirus).
Cell proliferation, EdU labeling, cell viability and colony formation assays
To measure cell proliferation, the cells were seeded in a 96-well plate at 1 × 104 per well. After seeding, the cell numbers were measured by MTT assay at the indicated days according to the manufacturer's instructions (Roche). Briefly, the MTT solution was added to the well and incubated for 4 h at 37 °C. The absorbance (490 nm) was measured and analyzed. In addition to MTT assays, the actual cell numbers from parallel experiments were counted at the indicated days. To measure DNA synthesis, EdU labeling was performed according to the manufacturer's instructions (Invitrogen). For cell viability assay, the cells were seeded at 1 × 104 per well. After 24 h, the cells were treated with different concentrations of doxorubicin or etoposide for another 18 h and followed by MTT assay. For colony-formation assay, 5 × 103 cells were seeded per well. MIF was added to medium 24 h after seeding when indicated. The culture media were changed every 2 days and supplemented with fresh MIF when indicated. The cells were allowed to grow for at least 2 weeks. To visualize the colony foci, the plates were fixed with 10% methanol and stained with 0.5% of crystal violet. DNA fragmentation assay was performed as described previously 72.
Transfection and luciferase reporter gene assay
Plasmid transfection using Lipofectamine reagents (Invitrogen) or Fugene HD (Roche) was carried out according to the manufacturers' instructions. After transfection (5 h), transfected cells were washed twice with phosphate-buffered saline, and fresh DMEM was added for further incubation. To determine the luciferase activity, whole-cell lysates were prepared and assayed as instructed by the manufacturer. The luciferase activity was normalized against total protein (Promega). Transfection of siRNAs was performed using the TransIT TKO reagents (Mirus).
Autophagy induction by GFP-LC3 punctate formation
MCF-7 cells stably expressing GFP-LC3 were obtained by transfecting the cells with pEGFP-LC3 plasmid and selected with G418. For autophagy induction, the formation of GFP-LC3 punctate structures was examined as previously described 73. The live cell images were taken using the Nikon Eclipse inverted fluorescent microscope, and the results were analyzed by the NES-Elements BR 3.10 software. Quantification of autophagy represented the ratio of GFP-LC3-punctate-positive cells to the total cells counted.
Tumor xenograft in athymic nude mice
Five-week-old female athymic nude mice (Hsd:Athymic Nude-nu; Harlan) were anesthetized with isoflurane before the injection. The cells used for injection (shControl and shMIF) were resuspended in DMEM-50% Matrigel at 107 cells per ml as described previously 61, 62. Each side of the #4 mammary gland fat pad was injected with of 106 cells (0.1 ml). The mice were monitored every week and the tumor sizes were measured with a caliper starting at 3 weeks after injection. The tumor volume was calculated by 1/6 × length × height × width × π. All animal experiments were approved by the Institutional Animal Care and Use Committee.
Immunoprecipitation and immunoblot analysis
Transfections were carried out essentially as described previously. The transfected cells were lysed in lysis buffer (20 mM Tris-HCl (pH 8.0), 125 mM NaCl, 0.5% NP-40, 2 mM EDTA, 0.2 mM NaF, 0.2 mM Na3VO4, protease inhibitor cocktail) for 30 min and the debris was cleared by centrifugation at 13 400× g for 20 min at 4 °C. For immunoprecipitation experiments, the lysates were incubated with 0.5 μg of anti-hemagglutinin (anti-HA; Roche Molecular Biochemicals) or anti-Flag (Sigma) antibody for 4 h or overnight at 4 °C. The antibody was allowed to bind to protein A and G beads for 1 h, and then washed extensively with lysis buffer. For western blot analysis, the samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). After blocking with 5% milk in TBST, the primary antibodies were diluted in TBST buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Tween 20) with 5% milk and added to the membranes for 1 h at room temperature (RT) or overnight at 4 °C, followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at RT. All blots were developed with Supersignal substrate (Pierce) and visualized by chemiluminescence. Subsequent probing with different antibodies was made possible by stripping the membranes with buffer (62.5 mM Tris-HCl (pH 6.8), 2% SDS, 100 mM β-mercaptoethanol) at 55 °C for 30 min.
Quantitative PCR analysis
The quantitative PCR was performed using the StepONE Plus real-time PCR system (Applied Biosystems). Primer sequences for the ChIP experiments are sense 5′-CCGCTCGAGTCCCCACTCGGGGCGGAGCCGCAG-3′ and anti-sense 5′- CCCAAGCTTGGCATGATGGCAGAAGGACCAGGAGACCC-3′.
RNA preparation, RNase protection assay and RT-PCR
Total RNA was prepared using the RNeasy kit according to the manufacturer's instructions (Qiagen). Total RNA of 5 μg was used in the RNase protection assay with the RNase protection assay kit (Pharmingen) and 2 μg were used for RT-PCR with the SuperScript III Platinum One-Step Quantitative RT-PCR system (Invitrogen).
Statistical analyses
All results are shown as the mean ± SD. The comparison of different groups was carried out by using two-tailed unpaired Student's t-test, and differences at or below P < 0.05 were considered statistically significant.
References
American Cancer Society. Available from: http://www.cancer.org/doctor/home/index.asp.
Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 1997; 277:965–968.
Cai D, Shames DS, Raso MG, et al. Steroid receptor coactivator-3 expression in lung cancer and its role in the regulation of cancer cell survival and proliferation. Cancer Res 2010; 70:6477–6485.
Dihge L, Bendahl PO, Grabau D, et al. Epidermal growth factor receptor (EGFR) and the estrogen receptor modulator amplified in breast cancer (AIB1) for predicting clinical outcome after adjuvant tamoxifen in breast cancer. Breast Cancer Res Treat 2008; 109:255–262.
Osborne CK, Bardou V, Hopp TA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst 2003; 95:353–361.
Chen HW, Lin RJ, Xie W, Wilpitz D, Evans RM . Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 1999; 98:675–686.
Feng Q, Yi P, Wong JM, O'Malley BW . Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 2006; 26:7846–7857.
Wu RC, Qin J, Yi P, et al. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic responses to multiple cellular signaling pathways. Mol Cell 2004; 15:937–949.
Conroy H, Mawhinney L, Donnelly SC . Inflammation and cancer: macrophage migration inhibitory factor (MIF)–the potential missing link. QJM 2010; 103:831–836.
Mitchell RA, Liao H, Chesney J, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci USA 2002; 99:345–350.
Nishihira JUN, Ishibashi T, Fukushima T, et al. Macrophage migration inhibitory factor (MIF). Ann NY Acad Sci 2003; 995:171–182.
Bach J-P, Deuster O, Balzer-Geldsetzer M, et al. The role of macrophage inhibitory factor in tumorigenesis and central nervous system tumors. Cancer 2009; 115:2031–2040.
Bach JP, Rinn B, Meyer B, Dodel R, Bacher M . Role of MIF in inflammation and tumorigenesis. Oncology 2008; 75:127–133.
del Vecchio MT, Tripodi SA, Arcuri F, et al. Macrophage migration inhibitory factor in prostatic adenocarcinoma: correlation with tumor grading and combination endocrine treatment-related changes. Prostate 2000; 45:51–57.
Markert JM, Fuller CM, Gillespie GY, et al. Differential gene expression profiling in human brain tumors. Physiol Genomics 2001; 5:21–33.
Meyer-Siegler KL, Bellino MA, Tannenbaum M . Macrophage migration inhibitory factor evaluation compared with prostate specific antigen as a biomarker in patients with prostate carcinoma. Cancer 2002; 94:1449–1456.
Winner M, Koong AC, Rendon BE, Zundel W, Mitchell RA . Amplification of tumor hypoxic responses by macrophage migration inhibitory factor-dependent hypoxia-inducible factor stabilization. Cancer Res 2007; 67:186–193.
Welford SM, Bedogni B, Gradin K, et al. HIF1alpha delays premature senescence through the activation of MIF. Genes Dev 2006; 20:3366–3371.
Wei N, Deng XW . The COP9 signalosome. Annu Rev Cell Dev Biol 2003; 19:261–286.
Wei N, Serino G, Deng XW . The COP9 signalosome: more than a protease. Trends Biochem Sci 2008; 33:592–600.
Chauchereau A, Georgiakaki M, Perrin-Wolff M, Milgrom E, Loosfelt H . JAB1 interacts with both the progesterone receptor and SRC-1. J Biol Chem 2000; 275:8540–8548.
Kleemann R, Hausser A, Geiger G, et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature 2000; 408:211–216.
Lue H, Kapurniotu A, Fingerle-Rowson G, et al. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal 2006; 18:688–703.
Lue H, Dewor M, Leng L, Bucala R, Bernhagen J . Activation of the JNK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on CXCR4 and CD74. Cell Signal 2011; 23:135–144.
Salminen A, Kaarniranta K . Control of p53 and NF-kappaB signaling by WIP1 and MIF: role in cellular senescence and organismal aging. Cell Signal 2011; 23:747–752.
Levine B, Klionsky DJ . Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463–477.
Mizushima N . The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ 2005; 12(Suppl 2):1535–1541.
Arico S, Petiot A, Bauvy C, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem 2001; 276:35243–35246.
Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A . Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev 1995; 9:15–30.
Futreal PA, Soderkvist P, Marks JR, et al. Detection of frequent allelic loss on proximal chromosome 17q in sporadic breast carcinoma using microsatellite length polymorphisms. Cancer Res 1992; 52:2624–2627.
Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809–1820.
Saito H, Inazawa J, Saito S, et al. Detailed deletion mapping of chromosome 17q in ovarian and breast cancers: 2-cM region on 17q21.3 often and commonly deleted in tumors. Cancer Res 1993; 53:3382–3385.
Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927–939.
Kitada S, Takayama S, De Riel K, Tanaka S, Reed JC . Reversal of chemoresistance of lymphoma cells by antisense-mediated reduction of bcl-2 gene expression. Antisense Res Dev 1994; 4:71–79.
Reed JC, Cuddy M, Slabiak T, Croce CM, Nowell PC . Oncogenic potential of bcl-2 demonstrated by gene transfer. Nature 1988; 336:259–261.
Tsujimoto Y, Cossman J, Jaffe E, Croce CM . Involvement of the bcl-2 gene in human follicular lymphoma. Science 1985; 228:1440–1443.
Zhang Y, Lu H . Signaling to p53: ribosomal proteins find their way. Cancer Cell 2009; 16:369–377.
Amazit L, Pasini L, Szafran AT, et al. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol Cell Biol 2007; 27:6913–6932.
York B, Yu C, Sagen JV, et al. Reprogramming the posttranslational code of SRC-3 confers a switch in mammalian systems biology. Proc Natl Acad Sci USA 2010; 107:11122–11127.
Park KJ, Krishnan V, O'Malley BW, Yamamoto Y, Gaynor RB . Formation of an IKKalpha-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol Cell 2005; 18:71–82.
Wu RC, Qin J, Hashimoto Y, et al. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by I kappa B kinase. Mol Cell Biol 2002; 22:3549–3561.
Schödel J, Oikonomopoulos S, Ragoussis J, et al. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 2011; 117:e207–e217.
Baugh JA, Gantier M, Li L, et al. Dual regulation of macrophage migration inhibitory factor (MIF) expression in hypoxia by CREB and HIF-1. Biochem Biophys Res Commun 2006; 347:895–903.
He H, Dang Y, Dai F, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 2003; 278:29278–29287.
Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488–492.
Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720–5728.
Kabeya Y, Mizushima N, Yamamoto A, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004; 117:2805–2812.
Tanida I, Ueno T, Kominami E . Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem 2004; 279:47704–47710.
Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD . Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected]. Nat Cell Biol 2006; 8:398–406.
Fung C, Lock R, Gao S, Salas E, Debnath J . Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 2008; 19:797–806.
Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 2001; 152:657–668.
Bjørkøy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on Huntingtin-induced cell death. J Cell Biol 2005; 171:603–614.
Bjørkøy G, Lamark T, Johansen T . p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2006; 2:138–139.
Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131:1149–1163.
Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672–676.
Akar U, Chaves-Reyez A, Barria M, et al. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy 2008; 4:669–679.
Choi CH, Jung YK, Oh SH . Autophagy induction by capsaicin in malignant human breast cells is modulated by p38 and extracellular signal-regulated mitogen-activated protein kinases and retards cell death by suppressing endoplasmic reticulum stress-mediated apoptosis. Mol Pharmacol 2010; 78:114–125.
Esteve JM, Armengod ME, Knecht E . BRCA1 negatively regulates formation of autophagic vacuoles in MCF-7 breast cancer cells. Exp Cell Res 2010; 316:2618–2629.
Hwang J, Kim H, Kim J, et al. Zinc(II) ion mediates tamoxifen-induced autophagy and cell death in MCF-7 breast cancer cell line. Biometals 2010; 23:997–1013.
Morton CL, Houghton PJ . Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc 2007; 2:247–250.
Yue W, Brodie A . MCF-7 human breast carcinomas in nude mice as a model for evaluating aromatase inhibitors. J Steroid Biochem Mol Biol 1993; 44:671–673.
Yue W, Zhou D, Chen S, Brodie A . A new nude mouse model for postmenopausal breast cancer using MCF-7 cells transfected with the human aromatase gene. Cancer Res 1994; 54:5092–5095.
Torres-Arzayus MI, de Mora JF, Yuan J, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell 2004; 6:263–274.
Torres-Arzayus MI, Zhao J, Bronson R, Brown M . Estrogen-dependent and estrogen-independent mechanisms contribute to AIB1-mediated tumor formation. Cancer Res 2010; 70:4102–4111.
Bucala R, Donnelly SC . Macrophage migration inhibitory factor: a probable link between inflammation and cancer. Immunity 2007; 26:281–285.
Lanz RB, Bulynko Y, Malovannaya A, et al. Global characterization of transcriptional impact of the SRC-3 coregulator. Mol Endocrinol 2010; 24:859–872.
Chi KC, Wallis AE, Lee CH, et al. Effects of Bcl-2 modulation with G3139 antisense oligonucleotide on human breast cancer cells are independent of inherent Bcl-2 protein expression. Breast Cancer Res Treat 2000; 63:199–212.
Lima RT, Martins LM, Guimaraes JE, Sambade C, Vasconcelos MH . Specific downregulation of bcl-2 and xIAP by RNAi enhances the effects of chemotherapeutic agents in MCF-7 human breast cancer cells. Cancer Gene Ther 2004; 11:309–316.
Carrero P, Okamoto K, Coumailleau P, et al. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol Cell Biol 2000; 20:402–415.
Wang F, Zhang R, Wu X, Hankinson O . Roles of coactivators in hypoxic induction of the erythropoietin gene. PLoS One 2010; 5:e10002.
Kuang SQ, Liao L, Zhang H, et al. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res 2004; 64:1875–1885.
Iwamoto K, Shinomiya N, Mochizuki H . Different cell cycle mechanisms between UV-induced and X-ray-induced apoptosis in WiDr colorectal carcinoma cells. Apoptosis 1999; 4:59–66.
Kimura S, Fujita N, Noda T, Yoshimori T . Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol 2009; 452:1–12.
Acknowledgements
We thank Drs David Lonard, Goberdhan Dimri and Valerie W Hu for their helpful comments on this manuscript. This work was supported in part by grants from Dan L Duncan Cancer Center, Baylor College of Medicine and Susan G Komen for the Cure (BCTR0707225 to RCW) and NIH (R01 CA119689 to JX).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Knockdown of SRC-3 or MIF inhibits cell proliferation. (PDF 377 kb)
Supplementary information, Figure S2
Cell cycle analysis. (PDF 74 kb)
Supplementary information, Figure S3
Activation of the MIF promoter by SRC-3 and not SRC-1 or SRC-2. (PDF 110 kb)
Supplementary information, Figure S4
Activation of the MIF promoter by HIF-1α requires the HRE. (PDF 144 kb)
Supplementary information, Figure S5
Synergistic activation of the MIF promoter by SRC-3 and HIF-1α. (PDF 63 kb)
Supplementary information, Figure S6
Recruitment of HIF-1α, SRC-3, or CBP to the endogenous MIF promoter. (PDF 112 kb)
Supplementary information, Figure S7
Knockdown of HIF-1α, SRC-3, or CBP by specific siRNAs. (PDF 65 kb)
Supplementary information, Figure S8
Knockdown of SRC-3 or MIF did not increase PARP cleavage or DNA fragmentation. (PDF 434 kb)
Supplementary information, Figure S9
Treatment of rapamycin induced punctuated GFP-LC3 structure. (PDF 238 kb)
Supplementary information, Figure S10
Expression of Beclin 1 in MCF-7 cells. (PDF 48 kb)
Supplementary information, Figure S11
Activation of ERK by MIF. (PDF 296 kb)
Supplementary information, Figure S12
Over-expression of MIF reduces chemosensitivity. (PDF 363 kb)
Supplementary information, Figure S13
Treatment with doxorubicin or etoposide did not increase PARP cleavage or DNA fragmentation in the MIF knockdown cells. (PDF 120 kb)
Rights and permissions
About this article
Cite this article
Wu, MY., Fu, J., Xu, J. et al. Steroid receptor coactivator 3 regulates autophagy in breast cancer cells through macrophage migration inhibitory factor. Cell Res 22, 1003–1021 (2012). https://doi.org/10.1038/cr.2012.44
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2012.44