Abstract
The genomes of myeloid malignancies are characterized by epigenomic abnormalities. Heterozygous, inactivating ten-eleven translocation 2 (TET2) mutations and neomorphic isocitrate dehydrogenase (IDH) mutations are recurrent and mutually exclusive in acute myeloid leukaemia genomes. Ascorbic acid (vitamin C) has been shown to stimulate the catalytic activity of TET2 in vitro and thus we sought to explore its effect in a leukaemic model expressing IDH1R132H. Vitamin C treatment induced an IDH1R132H-dependent reduction in cell proliferation and an increase in expression of genes involved in leukocyte differentiation. Vitamin C induced differentially methylated regions that displayed a significant overlap with enhancers implicated in myeloid differentiation and were enriched in sequence elements for the haematopoietic transcription factors CEBPβ, HIF1α, RUNX1 and PU.1. Chromatin immunoprecipitation sequencing of PU.1 and RUNX1 revealed a significant loss of PU.1 and increase of RUNX1-bound DNA elements accompanied by their demethylation following vitamin C treatment. In addition, vitamin C induced an increase in H3K27ac flanking sites bound by RUNX1. On the basis of these data we propose a model of vitamin C-induced epigenetic remodelling of transcription factor-binding sites driving differentiation in a leukaemic model.
Similar content being viewed by others
Introduction
Efforts to characterize genetic lesions in haematopoietic malignancies have revealed recurrent gain and loss-of-function mutations to epigenetic modifiers. Genomes of acute myeloid leukaemia (AML) patients frequently harbour recurrent mutations in the coding sequences of the genes ten-eleven translocation 2 (TET2) and isocitrate dehydrogenase 1 and 2 (IDH1/2) at diagnosis (8.5% for TET2 and 19.5% for IDH1/2) with recent studies suggesting such mutations may be among ‘founder’ mutations that exist in pre-leukaemic cells.1 Both of these enzymes are reported to have a role in DNA methylation homeostasis via perturbation of an as yet not fully characterized DNA demethylation pathway. One unifying notion is that de novo neomorphic IDH mutations promote aberrant DNA methylation by increased production of 2-hydroxyglutarate (2HG),2 which inhibits the hydroxylation of 5-methylcytosine (5mC), a reaction catalysed by TET2. In support of this notion, multiple reports have shown that the R-2-hydroxyglutarate initiates leukaemogenesis in a reversible manner.3, 4 AML patient genomes also demonstrate mutual exclusivity in mutations to IDH and TET2; and TET2-mutant AMLs show an overlapping promoter hypermethylation signature with IDH1/2-mutant AMLs.5 Survival data associated with TET2 and IDH1/2 mutations in AML patients are inconsistent and no prognostic significance to these mutations has been conclusively established.6, 7
The homeobox protein HOXA9 contributes to haematopoiesis and is frequently overexpressed in human AMLs,8 notably in patients harbouring the IDH1R132H mutation.9 Immortalization of murine bone marrow cells by retroviral HOXA9 expression leads to the expansion of myeloid progenitors resistant to terminal differentiation into monocytes and granulocytes via granulocyte–macrophage colony-stimulating factor and interleukin-3,10, 11 and in vivo transplantation experiments have shown that the overexpression of HOXA9 dysregulates MAPK signalling and collaborates with IDH1R132H to induce a short-latency AML in mice.9
Ascorbic acid (vitamin C) is a water-soluble, essential nutrient and common medium supplement shown to enhance cellular proliferation.12 Intracellular vitamin C is highly concentrated in immune and brain cells where it maintains iron in the Fe(II) state, a requirement for the catalytic activity of 2-oxoglutarate-dependent dioxygenases.12 In addition to its requirement as a cofactor, in vitro studies have provided evidence of a physical interaction between vitamin C and the catalytic domain of TET2 that enhances the enzymatic oxidation of 5mC to 5-hydroxy-methylcytosine (5hmC).13 The biological significance of the epigenetic modulation induced by vitamin C is illustrated through its ability to improve induced pluripotent stem cell generation14 and induce a blastocyst-like state in mouse embryonic stem cell15 by promoting the demethylation of H3K9 and 5mC, respectively. Multiple lines of evidence have suggested that vitamin C, in its native form, is toxic in culture through the formation of extracellular H2O2(refs 16, 17) potentially masking its more biologically relevant epigenetic effects. 2-phosphate ascorbic acid (vitC) is an oxidatively stable form of vitamin C that does not contribute extracellular H2O2 formation and is converted to ascorbic acid during transport across the cell membrane.18
The potential dichotomous effects of vitamin C versus IDH1R132H on haematologically relevant epigenetic regulators led us to leverage HOXA9-immortalized bone marrow cells expressing IDH1R132H as a model to explore the phenotypic and epigenomic effects of vitamin C treatment in a leukaemic model. Here we provide a survey of the chromatin landscape of HOXA9-IDH1R132H AML and new findings supporting a role for vitamin C in facilitating the epigenetic remodelling that occurs during differentiation of haematopoietic progenitors.
Materials and methods
Retroviral vectors and preparation of mouse bone marrow cells
Retroviral vectors MSCV-HoxA9-PGKneo, pSF91-IRES-eGFP, pSF91-IDH1mut-IRES-eGFP and pSF91-IDH1wt-IRES-eGFP have been described previously.9 C57BL/6J mice were injected intraperitoneally with 5-fluorouracil (Medac, Hamburg, Germany) at a dose of 150 mg/kg. Five days later, bone marrow cells were collected and transduced first by co-cultivation with a HoxA9 viral producer cell line followed by either IDH1wt or IDH1mut viral producer GP+E86 cells.9 Cells were then sorted for green fluorescent protein (GFP) expression and maintained in Dulbecco’s modified Eagle’s medium (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies, Burlington, ON, Canada) and 6 ng/ml murine interleukin-3, 10 ng/ml human interleukin-6 and 20 ng/ml murine stem cell factor (Peprotech, Hamburg, Germany).
In vitro proliferation assay
Cells were plated at a density of 5 × 105 cells/ml in flat-bottom 24-well plates and incubated under light-protective conditions. Vitamin C in the form of l-ascorbic acid 2-phosphate sesqui-magnesium salt hydrate (vitC; A8960; Sigma-Aldrich, Steinheim, Germany) was added to the culture medium daily at a concentration of 100 μg/ml (0.345 mm). Live cells were counted at the specified days with trypan blue.
Lineage staining and fluorescence-activated cell sorting antibodies
Cells were plated at a density of 5 × 105 cells/ml in flat-bottom 6-well plates (5 ml/well) under light-protective conditions and vitC was added at the specified concentrations. Monoclonal antibodies for lineage staining used were Gr1-PE (clone-RB6-8CS), Sca1-PE (D7), ckit-APC (2B8; BD Biosciences, Heidelberg, Germany), CD11b-APC (M1/70) and F4/80-PE (BM8; eBioscience, Frankfurt, Germany). Lineage distribution was determined by fluorescence-activated cell sorting analysis (FACSCalibur, Becton Dickinson, Heidelberg, Germany) as previously described.9
RNA-sequencing
Briefly, RNA was extracted from untreated and vitamin C-treated (0.345 mm added daily) IDH1R132H cells 72 h after initial treatment and subjected to RNA-seq library construction according to standard operating procedures (http://www.epigenomes.ca/protocols-and-standards).
Immunofluorescence staining
Untreated and vitC-treated (0.345 mm; 12 h) IDH1R132H cells were fixed in 4% paraformaldehyde for 15 min. After washing three times with phosphate-buffered saline (PBS), cells were blocked with 5% fetal bovine serum in PBS+0.5% Tween 20 for 2 h at room temperature (20–25 °C). Primary antibody for 5hmC (1:100, Active Motif, Carlsbad, CA, USA) was diluted in blocking solution and incubated with cells overnight at 4 °C. Cells were then washed three times in PBS and incubated for 2 h at room temperature with secondary antibodies diluted in blocking solution. Cells were washed three times in PBS and mounted (ProLong Diamond Antifade Mountant with 4,6-diamidino-2-phenylindole, Molecular Probes, Eugene, OR, USA) before imaging with Alexa Fluor 568-conjugated goat anti-mouse (1:1 000, Life Technologies) secondary antibody.
Methylated and hydroxyl-methylated DNA immunoprecipitation sequencing
Genomic DNA was collected from untreated and vitamin C-treated (0.345 mm added daily) IDH1R132 cells 72 h after initial treatment and subjected to low-input methylated DNA immunoprecipitation sequencing (meDIP-seq) and hydroxyl-meDIP-seq (hmeDIP-seq) as previously described19 (Supplementary Methods).
Whole-genome bisulfite sequencing
Genomic DNA was collected from untreated and vitamin C-treated (0.345 mm added daily) IDH1R132H cells 72 h after initial treatment. Bisulfite conversion was performed using the MethylEdge Kit (Promega, Madison, WI, USA) according to the manufacturer recommendations and subjected to cDNA synthesis followed by Illumina (Hayward, CA, USA) library preparation (Supplementary Methods), pooling and sequencing on the HiSeq 2500 sequencing platforms (v3 chemistry).
Histone mark chromatin immunoprecipitation sequencing
Chromatin immunoprecipitation sequencing (ChIP-seq) was performed on chromatin collected from untreated and vitC-treated (0.345 mm added daily) IDH1R132H cells. Standard operating procedures for native ChIP-seq20 library construction (Supplementary Methods) are also available (http://www.epigenomes.ca/protocols-and-standards) or by request.
Transcription factor ChIP-seq
Formaldehyde crosslinked transcription factor ChIP-seq library construction was performed on chromatin collected from untreated and vitC-treated (0.345 mm added daily) IDH1R132 cells 72 h after initial treatment using RUNX1 (Abcam, Cambridge, MA, USA; ab23980) and PU.1 (SCBT, Dallas, TX, USA; sc-22805) antibodies as previously described21 with minor modifications (Supplementary Methods). Libraries were pooled on the NextSeq 500 sequencing platform.
Bioinformatics
See Supplementary Methods for detailed, library-specific computational methods.
Results
Vitamin C reduces cell proliferation and increases mature haematopoietic populations
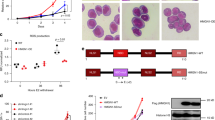
To explore the effect of vitamin C in a leukaemic context we utilized a previously established model in which murine bone marrow cells were immortalized by the overexpression of HOXA9(ref. 10) and subsequently transfected with a retroviral vector expressing either IDH1WTor IDH1R132H.9 Consistent with the notion that IDH1R132H inhibits TET activity through the production of 2-hydroxyglutarate (data not shown), IDH1R132H-expressing cells showed an overall reduction in 5hmC compared to IDH1WT cells (Figure 1a; Supplementary Figure S1a). We utilized 2-phosphate l-ascorbic acid (vitC), which, unlike native ascorbic acid, remains oxidatively stable under standard cell culture conditions.22 VitC treatment (0.345 mm; throughout unless otherwise indicated) induced a marked increase in 5hmC signal in IDH1R132H cells, but not in IDH1WT cells (Figure 1a; Supplementary Figure S1a). Daily vitC treatment over the course of 10 days induced a dose-dependent reduction in the proliferation rate of IDH1R132H cells and to a lesser extent IDH1WT cells (Figure 1b; Supplementary Figure S1b). Morphological assessment following vitC treatment revealed an increase in apoptosis (Supplementary Figure S1d) with the remaining viable cells differentiated into monocytes and macrophage cells (Supplementary Figure S1c) in IDH1R132H but not in IDH1wt cells. Further examination of cell phenotype by fluorescence-activated cell sorting reveals that vitC treatment increased the Mac1+F4/80+ (+13%) and Mac1+Gr1+ (+34%) populations and decreased the Sca1+ (−4%) and cKit+ (−30.8%) populations in IDH1R132H but not in IDH1WT cells (Figure 1c; Supplementary Figure S1e). Furthermore, colony-forming cell assays revealed a vitC-induced, dose-dependent reduction in colony-forming cells, an effect that was more pronounced in IDH1R132H compared to IDH1WT cells (Supplementary Figure S1f). Taken together these findings suggest that IDH1R132H promotes a more primitive phenotype and that vitC reduces proliferation and promotes the differentiation of myeloid progenitor cells in the context of our model.
Vitamin C reduces cell proliferation and modulates gene expression in IDH1R132H-expressing cells. (a) Mean fluorescence values from 5hmC immunofluorescence experiments±s.e.m. (b) A growth curve represented as mean cell count±s.d., for untreated (grey) or vitamin C-treated (orange; 0.345 mm) IDH1wt and IDH1R132H cells over 9 days. (c) The proportion of fluorescence-activated cell sorting sorted cells with each indicated marker alone or in combination. Untreated (grey) or vitamin C-treated (orange; 72 h) IDH1WT or IDH1R132H cells; represented by mean proportion±s.e.m. (d) RNA-seq expression values, represented as log10(RPKM), for each gene in mm10 Ensembl v71 (RPKM>0.01) in untreated (x axis) and vitamin C-treated (y axis) cells. Upregulated (up), downregulated (down) and unaffected genes (false discovery rate <0.05) are represented by orange, blue and black dots, respectively. (e) The top 10 GO results, represented by hyper-geometric fold enrichment, for upregulated genes upon vitamin C treatment. RPKM, reads per kilobase million.
Vitamin C induces the expression of a myeloid-specific gene signature
To investigate the molecular changes induced by vitC we performed mRNA-seq on IDH1R132H cells and observed 450 upregulated and 221 downregulated genes upon vitC treatment (Figure 1d). Upregulated genes were significantly enriched in Gene Ontology (GO) terms (47, Bonferroni P<0.01). The top enriched GO terms for upregulated genes included terms for positive regulation of leukocyte differentiation, chemotaxis and migration, negative regulation of cell proliferation (Figure 1e) and Kyoto Encyclopedia of Genes and Genomes terms for haematopoietic cell lineage, cytokine–cytokine interaction and Toll-like receptor signalling (Supplementary Figure S2a). In contrast, genes downregulated upon vitC treatment revealed no significantly enriched GO terms. Further examination revealed significant upregulation (false discovery rate <0.05) of multiple genes expressed in differentiated myeloid cells, including CSF2RA,23 ITGAM (Mac1), CEBPE24 and ELANE,25 and increased expression of genes encoding cell surface markers that define mature myeloid cells (Supplementary Figure S2b, upper). We validated the observed differential expression on a subset of genes by quantitative PCR (Supplementary Figure S2c).
The catalytic activity of all three TET family members (TET1, 2 and 3) are induced by vitC,13, 26 and both TET2 and TET3 are highly expressed in our model and in human AML27 (Supplementary Figure S2d). To determine the extent to which 5hmC gains and transcriptional alterations induced by vitC were dependent on TET2 activity we performed TET2 knockdown in IDH1R132H cells. TET2 knockdown was confirmed at the protein level by western blot analysis (Supplementary Figure S2e). As expected vitC treatment induced 5hmC gain in IDH1R132H cells treated with scrambled small interfering RNA but this effect was significantly reduced in cells treated with small interfering RNA targeting the TET2 transcript (Supplementary Figures S2f and g). To confirm whether the vitC-induced transcriptional alterations were dependent on TET2 we performed quantitative PCR on a set of genes upregulated following vitC treatment and observed reduced upregulation in all five targets tested (Supplementary Figure S2h; Supplementary Table 1). These results support a role for TET2 in mediating 5hmC gain and expression changes in response to vitC treatment.
Vitamin C modulates DNA methylation at genomic regions associated with myelopoiesis and AML
To explore qualitative changes in DNA methylation upon vitC treatment, genomic DNA was extracted from untreated and vitC-treated IDH1R132H and IDH1WT cells, and subjected to meDIP-seq or hmeDIP-seq, respectively. MACS28 was used to identify differentially methylated regions (DMRs; P<1e−4), and the MEDIPS R package29 was used to calculate significant changes (P<0.01) in meDIP-seq and hmeDIP-seq signal in 500 bp windows genome wide (Table 1). Genomic regions that lost 5mC in IDH1R132H cells upon vitC treatment (deMRs; 3789 regions) showed an increased 5hmC signal flanking the centre of deMRs (Figure 2a, top panel), a pattern consistent with the notion that TET-mediated hydroxylation of 5mC to 5hmC drives site-specific demethylation. We observed a similar 5hmC pattern flanking the centre of regions that gained methylation in IDH1R132H cells compared to IDH1WT cells (IDH1R132H iMRs; 4182 regions; Figure 2a, lower).
Vitamin C induces methylome remodelling in IDH1R132H-expressing cells. (a) Average normalized 5hmC (hmeDIP) signal for untreated (black) and vitamin C-treated (orange) IDH1R132H cells±2.5 kb from the centre of regions hypermethylated in IDH1R132H cells vs IDH1wt cells (IDH1R132H iMRs; top) and regions demethylated upon vitamin C treatment in IDH1R132H cells (deMRs; bottom). (b) Top five GREAT GO results for vitamin C deMRs (orange), iHRs (green) and the intersection between deMRs, iHRs and IDH1R132H iMRs (blue; n=23). (c) UCSC genome browser screenshot depicting meDIP, hmeDIP, whole-genome shotgun bisulfite sequencing (WGBS) and RNA-seq values at the Csf2ra promoter in both untreated (black) and vitamin C-treated (orange) IDH1R132H cells. (d) Top hits from HOMER de novo motif analysis performed on vitamin C deMRs identified by WGBS. (e) A heat map representing the proportion of regions in different DMR sets (y axis) that overlap mm10 genomic features (x axis) and transcription factor-binding sites from embryonic stem cells (esc; negative control), macrophages (myeloid) or haematopoietic progenitors (progenitor).
Having established specific qualitative differences in 5mC and 5hmC levels in IDH1R132H cells we sought to quantitatively assess 5mC changes in response to vitC by whole-genome shotgun bisulfite sequencing of genomic DNA extracted from untreated and vitC-treated IDH1R132H cells. We identified 6496 deMRs and 8374 iMRs (Table 1; Materials and Methods) induced by vitC in IDH1R132H cells. Consistent with the meDIP-seq results, intersection of whole-genome shotgun bisulfite sequencing vitC deMRs with hmeDIP-seq and meDIP-seq data revealed increased hmC and decreased mC levels within deMRs (Supplementary Figure S3a). A majority of vitC deMRs and iMRs were distal (>5 kb) to gene transcription start sites and iMRs were enriched at CpG islands (Supplementary Figure S3b).
Functional enrichment of genes associated with vitC deMRs30 and regions that gained 5hmC upon vitC treatment in IDH1R132H cells (vitC iHRs; n=7829) revealed statistically significant (q<0.01) enrichment for gene sets related to haematopoiesis (Figure 2b), including the MAPK (q=6.47e−8) and the CXCR4 (q=6.8e−6) signalling pathways and GO terms related to myeloid cell differentiation (q=5.7e−10). We identified a set of vitC deMRs that were within 1 kb of both an IDH1R132H iMR and a vitC iHR (deMR/iHR/mut iMR; n=23). Strikingly, these regions were found to be in association with genes annotated as downregulated in primitive haematopoietic cells overexpressing either NUP98–HOXA9 (q=0.003) or RUNX1–RUNX1T1 (AML1–ETO; q=0.009; Figure 2b, blue). To identify regions likely undergoing TET-dependent, active demethylation (via 5hmC) we defined a set of regions comprised of the union of deMRs identified by meDIP-seq and whole-genome shotgun bisulfite sequencing that were within 1.5 kb of an iHR (DxMRs; n=712; Table 1). Functional enrichment analysis of the genes associated with DxMRs using GREAT30 confirmed previously identified pathways with increased enrichment for genes downregulated upon expression of NUP98–HOXA9 (+3.7 hyperFE vs deMR alone) and AML–ETO9a (+1.64 hyperFE vs deMR alone). This pattern of co-occurring gain of 5hmC and loss of 5mC is exemplified at the promoter of CSF2RA, a gene that is involved in myeloid differentiation and upregulated upon vitC treatment (Figure 2c). Reduced fractional methylation following vitC treatment within the deMR at the CSF2RA promoter was confirmed by targeted bisulfite sequencing in biological triplicates, along with six other selected gene-associated deMRs (Supplementary Figure S3c; Supplementary Table 2).
Functional annotation of genes associated with vitC iMRs show little association to haematopoiesis but reveal various terms related to nervous system function and development (Supplementary Figure S3d). Gains in 5mC would not be expected to be a direct consequence of vitC-induced TET activation but rather the consequence of indirect action. Interestingly, MEIS1, a gene previously described to increase expression in the presence of IDH2R140Q,31 showed an increased promoter methylation and downregulated expression (1.8-fold) in response to vitC. GREAT30 functional enrichment analysis of genes with promoter CpG islands containing iMRs revealed an enrichment for regions marked by H3K27me3 and H3K4me3 in embryonic stem cells and the ‘adult tissue stem module’ (false discovery rate q=1.72e−40), which represents a set of genes that are coordinately upregulated in somatic stem and leukaemic cells, including MEIS1(ref. 32) and CDK1,33 both of which are downregulated by vitC. Together, these observations suggest that in IDH1R132H-expressing cells, vitC treatment leads to a specific loss of DNA methylation at regulatory regions implicated in the control of haematopoiesis, which are associated with genes that are repressed in AML and upregulated in mature myeloid populations.
Vitamin C induces demethylation at haematopoietic transcription factor-binding sites
DNA methylation has been shown to influence the binding affinities of transcription factors34 and thus we sought to examine the link between vitC deMRs and transcription factors by identifying enriched motifs within vitC deMRs. Consistent with the observed GO associations, the top de novo motifs identified by HOMER35 were for PU.1 (34%), CEBPβ (15%), HIF1α (68%) and RUNX1 (30%; Figure 2d). Restricting our analysis to DxMRs increased the proportion of regions with de novo motifs for PU.1 (41%) and CEBPβ (41%) motifs while the proportion containing a RUNX1 motif decreased to 5%. In contrast, no de novo motifs were present in >1% of our iMR regions, suggesting vitC-induced increases in methylation are not associated with transcription factor-binding motifs.
To further explore the relationship between vitC DMRs and transcription factor binding we intersected PU.1 and CEBPβ ChIP-seq peaks and DNase I hypersensitive sites (DHSs) identified in haematopoietic progenitors and macrophages,36 and included embryonic stem cell enhancers37 and NANOG38 and RAD21(ref. 39) transcription factor-binding sites as negative controls (Figure 2e). VitC deMRs significantly overlapped mature myeloid (macrophage) DHSs (1019; 16%) and PU.1 (1846; 28%)- and CEBPβ (1428; 22%)-binding sites individually, with 768 deMRs (12%) overlapping regions bound by both transcription factors in macrophages. Furthermore, the average 5mC level was decreased in response to vitC treatment at macrophage PU.1-binding sites (t-test; P=3.7e−162), CEBPβ-binding sites (t-test; P=1.35e−119) and DHS (t-test; P=7.95e−47), but not progenitor DHS (t-test; P=0.447; Supplementary Figure S3e). VitC iMRs shared minimal overlap with myeloid transcription factor binding sites (Figure 2e). A significant proportion (226; 31%) of iMR containing CpG islands overlap haematopoietic progenitor DHSs, 95% of which become condensed during differentiation to macrophages. These results support a model of vitC-dependent demethylation preferentially at binding sites of transcription factors implicated in myeloid development.
Vitamin C-induced demethylation is enriched at myeloid-specific enhancers
Dynamic epigenetic remodelling at enhancer regions is a crucial step in cell fate determination of haematopoietic progenitor cells, a process that is often disturbed in haematological malignancies. Hypermethylation at enhancer elements coincides with leukaemogenesis in haematopoietic progenitor cells of TET2−/− mice expressing the AML–ETO9a fusion.40 In addition, the presence of DNA methylation has been shown to negatively correlate with enhancer activity41 and alters the ability of tissue-specific DNA-binding proteins to bind their target sites.34 Enhancer activity, represented by H3K4me1 signal, becomes established at the root of lineage commitment and increases as lineage-specific progenitor cells mature. H3K4me1 signal at 48 415 haematopoietic enhancers in 16 unique cell types37 was leveraged to explore the relationship between vitC-induced demethylation and enhancer dynamics during haematopoiesis. Strikingly, intersection of 48 415 haematopoietic enhancers with vitC deMRs revealed a significant overlap (1584; 24.4%; t-test P~0), 43% of which contained PU.1-binding sites. Plotting the mean H3K4me1 signal at vitC deMR-containing enhancers during lineage commitment from short term haematopoietic stem cells (ST-HSC) to the myeloid granulocyte-monocyte, common lymphoid and megakaryocyte-erythrocyte progenitor lineage progenitors revealed an increase in H3K4me1 signal at deMR associated enhancers during the progression from ST-HSC towards committed myeloid precursors granulocyte-monocyte, but not common lymphoid or megakaryocyte-erythrocyte progenitor cells (Figure 3a). Enhancers overlapping iMRs became less active, losing H3K4me1 signal in committed progenitors of all three lineages (Figure 3a). In mature haematopoietic cell types H3K4me1 signal increased at vitC deMR enhancers (K–S test; p~0) and decreased at vitC iMR containing enhancers (K–S test; P=1.25e−12) compared to random enhancers (n=1000) in the myeloid lineage (Figure 3b and Supplementary Figure S3f). These results suggest that vitC-induced demethylation occurs at enhancers that become active as haematopoietic progenitor cells differentiate into mature cells of the myeloid lineage.
Vitamin C treatment leads to demethylation at enhancers that become active in the myeloid lineage specifically. A depiction of the hierarchical haematopoietic differentiation pathway from HSC into mature cells of the erythroid (top), myeloid (middle) and lymphoid (bottom) lineages; (a) a linear representation of mean H3K4me1 signal during lineage commitment; and (b) a boxplot representing log10(H3K4me1 signal) at enhancers in mature cell types of each lineage at previously identified enhancers overlapping deMRs (orange), iMRs (blue) or a random set (n=1000; grey). HSC, hematopoietic stem cells.
H3K4 methylation and H3K27ac changes at vitamin C DMRs
In addition to the absence of 5mC, the presence of H3K4me1 and H3K27ac are hallmarks of active enhancers. To explore changes in the distribution of histone modifications and enhancer activity upon vitC treatment we performed H3K27ac, H3K4me1 and H3K4me3 ChIP-seq on IDH1R132H cells grown in the presence and absence of vitC. We first examined the distribution of histone marks in 500 bp regions surrounding the centre of vitC-induced DMRs and observed an increase in H3K27ac at deMRs, iHRs and DxMRs (t-test; P<1.34e−9), but not at iMRs (t-test; P=0.472; Figure 4a). We also observed increased H3K4me1 at all DMRs (t-test; P<3.37e−6) and increased H3K4me3 signal at deMRs (t-test; P=1.34e−4) and DxMRs (t-test; P=0.03) but not at iHRs (t-test; P=0.42) or iMRs (t-test; P=0.81). This pattern of change in H3K4me1/me3 and H3K27ac occupancy was validated in an independent biological replicate (Supplementary Figure S3g). These results support the notion that deMRs, DxMRs and iHRs, unlike iMRs, overlap with regulatory elements that undergo multifaceted epigenetic remodelling during vitC treatment.
Vitamin C induces changes in histone methylation and acetylation at DMRs and enhancers. (a) Average normalized signal difference between treatments (vitC−untreated) for H3K27ac (left) H3K4me1 (middle) and H3K4me3 (right)±1.5 kb from the centre of vitamin C DMRs. (b) Average normalized H3K27ac signal in the nearest enhancers (within 20 kb) to differentially expressed genes with * indicating statistically significant (P<0.01) differences between untreated (grey) and vitamin C-treated (orange) samples. (c) Average fractional methylation for vitamin C-treated (orange) or untreated (grey) cells at CpG sites overlapping IDH1R132H enhancers identified only in vitamin C-treated (top) or untreated (bottom) IDH1R132H cells, and (d) a linear representation of the average change in H3K4me1 signal during lineage commitment. (e) A boxplot representing H3K4me1 signal in enhancers in mature cell types of a given lineage at previously identified enhancers that overlap IDH1R132H enhancers identified in vitamin C-treated (orange) or untreated (blue) IDH1R132H cells. Random enhancers are included as a control (n=1000; grey).
We next examined the link between changes in histone modifications and gene expression upon vitC treatment. The majority of up- (373/450; 83%) and downregulated (165/221; 75%) genes are associated with one or more haematopoietic enhancers37 within 20 kb of their transcription start sites. Enhancers associated with genes that were transcriptionally induced upon vitC treatment showed a significant increase in H3K27ac signal (K–S test; P=0.0086; Figure 4b), suggesting that increased enhancer activity, represented by the acquisition of H3K27ac, coincides with increased expression of nearby genes. We next identified enhancers within IDH1R132H cells as those regions marked by H3K4me1 and H3K27ac (MACS2; P<1e−5) in either untreated or vitC-treated cells, removing those present in both cell types. This produced a set of 4389 and 3650 enhancers in untreated and vitamin C-treated cells, respectively. We observed significant loss of 5mC (t-test; P=7.94e−5) at enhancers active in vitC-treated but not in those specific to untreated IDH1R132H cells (t-test; P=0.8357; Figure 4c). To explore the activity of IDH1R132H cell enhancers during haematopoiesis we intersected them with the catalogue of haematopoietic enhancers.37 A significant fraction of IDH1R132H cell enhancers overlapped the catalogue of haematopoietic enhancers (1358/4389 (30%) and 1276/3650 (35%) for untreated and vitC-treated, respectively). Interestingly, although the majority of vitC-specific enhancers did not show a vitC-responsive loss of 5mC (1138/1276; 89.2%), they displayed a pattern of activation similar to deMR-containing enhancers during linage myeloid commitment (Figure 4d, centre panel) and had higher H3K4me1 signal in mature myeloid cells compared to random regions (K–S test; P~0; Figure 4e). These results suggest that changes in histone modifications upon vitC treatment, like changes in DNA methylation, occur at regulatory elements that become activated during the myelopoiesis.
Vitamin C alters the binding patterns of PU.1 and RUNX1
PU.1 is a pioneering transcription factor that is essential for haematopoiesis and, unlike most transcription factors, can bind chromatin in the presence of restrictive epigenetic modifications.42 Reduced PU.1 expression in haematopoietic cells leads to reduced growth and transformative capability in several leukaemic mouse models including AML.43 In addition, PU.1 has been implicated in the control of the expression of several genes that were differentially expressed upon vitC treatment (for example, KIT, CSF1 and CSF2RA). RUNX1 is a critical regulator of haematopoiesis that frequently participates in leukaemia-associated gene aberrations, including the RUNX1–RUNX1T1 (AML1–ETO9a) fusion recurrent in myeloid malignancies.44
To explore the effect of vitC treatment on the binding of these key haematopoietic transcription factors we performed ChIP-seq using antibodies against RUNX1 and PU.1 (transcription factor-ChIP) on IDH1R132H cells grown in the presence or absence of vitC. We identified enriched regions using MACS2(ref. 28) (narrow peaks; P<1e−5) and observed 62 770 and 23 781 PU.1-enriched regions in untreated and vitC-treated cells, respectively. Interestingly, the majority of PU.1-binding sites in vitC-treated cells were also found in untreated cells (22 646/23 781; 95%) suggesting a significant loss of PU.1 binding upon vitC treatment. We found that a significant proportion of vitC DMRs (30%; Figure 5a) and IDH1R132H enhancers (1252/3650, 34% vitC; and 1651/4389, 38% untreated) were located within 1 kb of PU.1-binding sites. These results suggest that PU.1 may guide the epigenomic remodelling events that occur upon vitC treatment.
Vitamin C-dependent changes in PU.1 and RUNX1 DNA binding and associated epigenetic states. (a) The proportion of different DMR regions within 1 kb of at least one PU.1 site identified by ChIP-seq. (b) Average (smoothed) fractional methylation for untreated (black) and vitamin C-treated (orange) cells at CpG sites±1.5 kb from the centre of vitamin C-specific RUNX1-binding sites that overlapped either CpG islands (left) or enhancers (right). * indicates a statistically significant difference (K–S test; P<0.05) between treatments. (c) Mean normalized H3K27ac (left), H3K4me1 (middle) and H3K4me3 (right) signal difference (vitC−untreated)±2.5 kb from the centre of regions flanking the summit of untreated-only (green) or vitamin C-only (blue) RUNX1-binding sites. (d) A proposed model of vitamin C-induced epigenetic remodelling.
We identified 4047 and 6587 RUNX1-enriched regions in untreated and vitC-treated cells, respectively. In contrast to PU.1, a large fraction of RUNX1-binding sites were specific to vitC-treated cells (3134/6587; 48%). A majority of vitamin C-specific RUNX1-binding events were within CpG islands (2128/3134; 68%) with a subset localizing to IDH1R132H cell enhancers (214/3134; 7%). Examining DNA methylation values within vitC-specific RUNX1-enriched regions revealed significant demethylation flanking RUNX1-enriched summits (±100 bp) within enhancers (K–S test; P=0.0024), but not at CpG islands (K–S test; P=0.2372; Figure 5b).
RUNX1 acts in concert with epigenetic modifiers, most notably histone acetyl-transferases, and perturbation of this ability can facilitate transformation.45 We examined the relationship between H3K27ac and H3K4me1 and vitC-induced RUNX1 binding by plotting the difference in signal in the presence and absence of vitC (vitC−untreated). VitC-specific RUNX1-enriched regions showed a significant gain in H3K4me1 and H3K27ac signal (K–S test; P~0) in the 500 bp flanking the centre of RUNX1-binding sites (Figure 5c). In contrast, we observed a significant loss of H3K4me1/me3 (K–S test; P~0), and to a lesser extent H3K27ac signal (K–S test; P=1.626e−6), upon vitC treatment in the region ±500 bp from the centre of RUNX1-enriched regions lost in response to vitC treatment (Figure 5c). This pattern of change in H3K4me1/me3 and H3K27ac relative to the position of each type of RUNX1-binding site was validated in an independent biological replicate (Supplementary Figure S3h). Taken together these results suggest that vitC treatment increases RUNX1 binding and increases H3K27ac and H3K4me1 in the flanking genomic regions.
Discussion
In this study we present evidence supporting a model of vitC-induced epigenomic remodelling that coincides with the differentiation and maturation of myeloid progenitor cells, a process frequently disrupted in myeloid malignancies. Using a murine leukaemic model expressing IDH1R132H we show that vitC treatment promotes DNA demethylation and H3K4me1 deposition at enhancers implicated in myeloid differentiation. We show that regions demethylated upon vitC treatment are enriched in putative regulatory elements associated with genes implicated in haematopoiesis and leukaemic transformation, and describe several key haematopoietic genes that show increased expression and promoter demethylation. We observe significant overlap between vitC-induced DMRs and PU.1-binding sites, consistent with previously described physical association between PU.1 and TET2.46 These observations support a model in which PU.1 guides TET2 to a specific subset of genomic regions to facilitate the oxidation of 5mC to 5hmC, a reaction that is inhibited by 2-hydroxyglutarate47 and enhanced by vitamin C13 (Figure 5d). Although demethylation events were observed in association with PU.1 sites, PU.1 binding did not increase upon vitC treatment, consistent with the ability of PU.1 to bind 5mC, unlike other E-twenty-six family members.48
The oncogenic t(8;21) AML–ETO9a translocation is recurrent in AML and fuses RUNX1 to RUNX1T1 leading to aberrant HDAC2 recruitment and histone deacetylation.49 We observe a relationship between vitC-induced DMRs and genes known to be dysregulated by this fusion protein and show that H3K27ac and H3K4me1 signal increases in the regions flanking vitC-induced RUNX1-binding sites. Interestingly, many regions that acquire H3K4me1 and H3K27ac upon vitC treatment overlap with previously described enhancers that become activated during myeloid development, but are devoid of deMRs. This suggests that vitC elicits multifaceted, myeloid-specific, epigenomic reprogramming that is not restricted to changes in 5mC and 5hmC.
While observational studies using oral and intravenous vitamin C suggested therapeutic benefit for a subset of cancer patients50, 51, 52 subsequent randomized, placebo-controlled trials revealed insignificant efficacy.53, 54 However, these latter studies utilized oral vitamin C, which induces transient micromolar increases in plasma levels55 significantly below the concentration used in our study (0.345 mm). Interestingly, a recent report in a small cohort of patients (n=24) with haematological malignancies showed that >90% had decreased plasma vitamin C concentrations and 58% were classified as vitamin C deficient.56 In contrast, only 7.1% of the general US population are vitamin C deficient.57
Our study provides mechanistic insight into a ‘first in class’ epigenetic modulator capable of re-activating an epigenetic pathway recurrently disrupted in cancer. This mechanistic insight, combined with our refined understanding of the role of epigenetic disruption in leukaemia, suggests that further investigation of the effects of millimolar concentrations of vitamin C in AML patients harbouring IDH and TET2 mutations, achievable in plasma through intravenous administration,58 is warranted. Indeed the lack of an appropriate, genotypically stratified cohort of patients may have contributed to the previously observed patient dependent response to vitamin C treatment.
References
Chan SM, Majeti R . Role of DNMT3A, TET2, and IDH1/2 mutations in pre-leukemic stem cells in acute myeloid leukemia. Int J Hematol 2013; 98: 648–657.
Losman J, Kaelin WG . What a difference a hydroxyl makes: mutant IDH,(R)-2-hydroxyglutarate, and cancer. Genes Dev 2013; 27: 836–852.
Chaturvedi A, Cruz MA, Jyotsana N, Sharma A, Goparaju R, Schwarzer A et al. Enantiomer-specific and paracrine leukemogenicity of mutant IDH metabolite 2-hydroxyglutarate. Leukemia 2016; 30: 1708–1715.
Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013; 339: 1621–1625.
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567.
Gaidzik VI, Paschka P, Spath D, Habdank M, Kohne CH, Germing U et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol 2012; 30: 1350–1357.
Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood 2010; 116: 2122–2126.
Argiropoulos B, Humphries R . Hox genes in hematopoiesis and leukemogenesis. Oncogene 2007; 26: 6766–6776.
Chaturvedi A, Cruz MMA, Jyotsana N, Sharma A, Yun H, Grlich K et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood 2013; 122: 2877–2887.
Calvo KR, Sykes DB, Pasillas M, Kamps MP . Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol Cell Biol 2000; 20: 3274–3285.
Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 2002; 99: 121–129.
Monfort A, Wutz A . Breathing-in epigenetic change with vitamin C. EMBO Rep 2013; 14: 337–346.
Yin R, Mao S, Zhao B, Chong Z, Yang Y, Zhao C et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc 2013; 135: 10396–10403.
Chen J, Liu H, Liu J, Qi J, Wei B, Yang J et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet 2013; 45: 34–42.
Blaschke K, Ebata KT, Karimi MM, Zepeda-Martnez JA, Goyal P, Mahapatra S et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 2013; 500: 222–226.
Chen Q, Espey MG, Krishna MC, Mitchell JB, Corpe CP, Buettner GR et al. Pharmacologic ascorbic acid concentrations selectively kill cancer cells: action as a pro-drug to deliver hydrogen peroxide to tissues. Proc Natl Acad Sci USA 2005; 102: 13604–13609.
Kawada H, Kaneko M, Sawanobori M, Uno T, Matsuzawa H, Nakamura Y et al. High concentrations of L-ascorbic acid specifically inhibit the growth of human leukemic cells via downregulation of HIF-1α transcription. PLoS One 2013; 8: e62717.
Takamizawa S, Maehata Y, Imai K, Senoo H, Sato S, Hata R . Effects of ascorbic acid and ascorbic acid 2-phosphate, a long-acting vitamin C derivative, on the proliferation and differentiation of human osteoblast-like cells. Cell Biol Int 2004; 28: 255–265.
Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D et al. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc 2012; 7: 617–636.
Lorzadeh A, Bilenky M, Hammond C, Knapp DJ, Li L, Miller PH et al. Nucleosome density ChIP-Seq identifies distinct chromatin modification signatures associated with MNase accessibility. Cell Rep 2016; 17: 2112–2124.
Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT . ChIP-seq: using high-throughput sequencing to discover protein–DNA interactions. Methods 2009; 48: 240–248.
Austria R, Semenzato A, Bettero A . Stability of vitamin C derivatives in solution and topical formulations. J Pharm Biomed Anal 1997; 15: 795–801.
de Groot RP, Coffer PJ, Koenderman L . Regulation of proliferation, differentiation and survival by the IL-3/IL-5/GM-CSF receptor family. Cell Signal 1998; 10: 619–628.
Tavor S, Vuong PT, Park DJ, Gombart AF, Cohen AH, Koeffler HP . Macrophage functional maturation and cytokine production are impaired in C/EBP epsilon-deficient mice. Blood 2002; 99: 1794–1801.
Wang Q, Li N, Wang X, Shen J, Hong X, Yu H et al. Membrane protein hMYADM preferentially expressed in myeloid cells is up-regulated during differentiation of stem cells and myeloid leukemia cells. Life Sci 2007; 80: 420–429.
Yue X, Trifari S, Aijo T, Tsagaratou A, Pastor WA, Zepeda-Martinez JA et al. Control of Foxp3 stability through modulation of TET activity. J Exp Med 2016; 213: 377–397.
Network CGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: 1.
Lienhard M, Grimm C, Morkel M, Herwig R, Chavez L . MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014; 30: 284–286.
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28: 495–501.
Ogawara Y, Katsumoto T, Aikawa Y, Shima Y, Kagiyama Y, Soga T et al. IDH2 and NPM1 mutations cooperate to activate Hoxa9/Meis1 and hypoxia pathways in acute myeloid leukemia. Cancer Res 2015; 75: 2005–2016.
Argiropoulos B, Yung E, Humphries RK . Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev 2007; 21: 2845–2849.
Radomska HS, Alberich-Jorda M, Will B, Gonzalez D, Delwel R, Tenen DG . Targeting CDK1 promotes FLT3-activated acute myeloid leukemia differentiation through C/EBPalpha. J Clin Invest 2012; 122: 2955–2966.
Domcke S, Bardet AF, Ginno PA, Hartl D, Burger L, Schbeler D . Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 2015; 528: 575–579.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589.
Goode DK, Obier N, Vijayabaskar M, Lie-A-Ling M, Lilly AJ, Hannah R et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev Cell 2016; 36: 572–587.
Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E et al. Chromatin state dynamics during blood formation. Science 2014; 345: 943–949.
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106–1117.
Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H et al. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One 2011; 6: e19470.
Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev 2015; 29: 910–922.
Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J 2011; 30: 3028–3039.
Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA 2008; 105: 6057–6062.
Zhou J, Wu J, Li B, Liu D, Yu J, Yan X et al. PU. 1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia 2014; 28: 1436–48v.
Okuda T, Nishimura M, Nakao M, Fujitaa Y . RUNX1/AML1: a central player in hematopoiesis. Int J Hematol 2001; 74: 252–257.
Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011; 333: 765–769.
de la Rica L, Rodríguez-Ubreva J, García M, Islam AB, Urquiza JM, Hernando H et al. PU. 1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol 2013; 14: 1.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19: 17–30.
Stephens DC, Poon GM . Differential sensitivity to methylated DNA by ETS-family transcription factors is intrinsically encoded in their DNA-binding domains. Nucleic Acids Res 2016; 44: 8671–8681.
Hu Z, Gu X, Baraoidan K, Ibanez V, Sharma A, Kadkol S et al. RUNX1 regulates corepressor interactions of PU.1. Blood 2011; 117: 6498–6508.
Cameron E, Campbell A . The orthomolecular treatment of cancer II. Clinical trial of high-dose ascorbic acid supplements in advanced human cancer. Chem Biol Interact 1974; 9: 285–315.
Cameron E, Pauling L . Supplemental ascorbate in the supportive treatment of cancer: prolongation of survival times in terminal human cancer. Proc Natl Acad Sci USA 1976; 73: 3685–3689.
Cameron E, Pauling L . Supplemental ascorbate in the supportive treatment of cancer: reevaluation of prolongation of survival times in terminal human cancer. Proc Natl Acad Sci USA 1978; 75: 4538–4542.
Creagan ET, Moertel CG, O'Fallon JR, Schutt AJ, O'Connell MJ, Rubin J et al. Failure of high-dose vitamin C (ascorbic acid) therapy to benefit patients with advanced cancer: a controlled trial. N Engl J Med 1979; 301: 687–690.
Moertel CG, Fleming TR, Creagan ET, Rubin J, O'Connell MJ, Ames MM . High-dose vitamin C versus placebo in the treatment of patients with advanced cancer who have had no prior chemotherapy: a randomized double-blind comparison. N Engl J Med 1985; 312: 137–141.
Levine M, Padayatty SJ, Espey MG . Vitamin C: a concentration-function approach yields pharmacology and therapeutic discoveries. Adv Nutr 2011; 2: 78–88.
Liu M, Ohtani H, Zhou W, Orskov AD, Charlet J, Zhang YW et al. Vitamin C increases viral mimicry induced by 5-aza-2'-deoxycytidine. Proc Natl Acad Sci USA 2016; 113: 10238–10244.
Schleicher RL, Carroll MD, Ford ES, Lacher DA . Serum vitamin C and the prevalence of vitamin C deficiency in the United States: 2003-2004 National Health and Nutrition Examination Survey (NHANES). Am J Clin Nutr 2009; 90: 1252–1263.
Padayatty SJ, Sun H, Wang Y, Riordan HD, Hewitt SM, Katz A et al. Vitamin C pharmacokinetics: implications for oral and intravenous use. Ann Intern Med 2004; 140: 533–537.
Acknowledgements
This work was supported by grants from the Terry Fox Research Institute Program Project (Grant: TFRI #1039) to MH and RKH, the Canadian Cancer Society Research Institute (Grant: CCSRI #703489) to MH, and grants 70111267 from Deutsche Krebshilfe to M Heuser (DFG grants HE 5240/5-1 and HE 5240/6-1). Sequencing data sets are available under GEO series GSE87623.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Mingay, M., Chaturvedi, A., Bilenky, M. et al. Vitamin C-induced epigenomic remodelling in IDH1 mutant acute myeloid leukaemia. Leukemia 32, 11–20 (2018). https://doi.org/10.1038/leu.2017.171
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.171
This article is cited by
Polycomb contraction differentially regulates terminal human hematopoietic differentiation programs
BMC Biology (2022)
Role of TET dioxygenases in the regulation of both normal and pathological hematopoiesis
Journal of Experimental & Clinical Cancer Research (2022)
High-dose intravenous vitamin C, a promising multi-targeting agent in the treatment of cancer
Journal of Experimental & Clinical Cancer Research (2021)
Differentiation therapy for myeloid malignancies: beyond cytotoxicity
Blood Cancer Journal (2021)
Context dependent effects of ascorbic acid treatment in TET2 mutant myeloid neoplasia
Communications Biology (2020)