Abstract
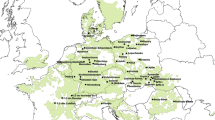
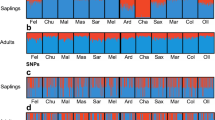
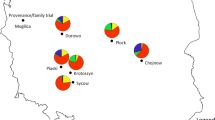
Although European beech (Fagus sylvatica L.) is one of the most widespread and ecologically and commercially most important deciduous trees in Europe, little is known about its adaptive genetic variation. We explored single-nucleotide polymorphism (SNP) variation in candidate genes for budburst and drought resistance in beech populations sampled across most of the distribution range, represented in an international provenance trial. SNP variation was monitored for six candidate genes, in 114 individuals from 19 natural populations. Population structure was deduced from the analysis of 20 nuclear microsatellite markers. Different methods to detect imprints of natural selection were used (F ST-outlier, SNP-climate regression, association tests). The F ST-outlier approach identified the COV gene with unambiguous signal of selection, which is an orthologue of Arabidopsis gene for a membrane protein previously reported as phenology-related. Based on environmental association analysis at the population level, the dehydrin gene was found associated with drought-related climatic variables. At the individual level, dehydrin gene also showed a significant association with chlorophyll fluorescence parameters, which are considered stress markers. The importance of the knowledge of physiological variation and geographical patterns of adaptive genetic variation for guiding reproductive materials transfer under climate change is stressed.
Similar content being viewed by others
References
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G (2008) LOSITAN: a workbench to detect molecular adaptation based on a F ST-outlier method. BMC Bioinformatics 9:323. doi:10.1186/1471-2105-9-323
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B 263:1619–1626
Bergmann F, Gregorius H-R (1993) Ecogeographical distribution and thermostability of isocitrate dehydrogenase (IDH) alloenzymes in European silver fir (Abies alba). Biochem Syst Ecol 21:597–605
Bošeľa M, Popa I, Gömöry D, Longauer R, Tobin B, Kyncl J, Kyncl T, Nechita C, Petráš R, Sidor C, Šebeň V, Büntgen U (2016) Effects of post-glacial phylogeny and genetic diversity on the growth variability and climate sensitivity of European silver fir. J Ecol 104:716–724
Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Briggs D, Walters SM (1997) Plant variation and evolution, 3rd edn. Cambridge University Press, Cambridge, 521 + xxi pp
Chen J, Källman T, Ma X, Gyllenstrand N, Zaina G, Morgante M, Bousquet J, Eckert A, Wegrzyn J, Neale D, Lagercrantz U, Lascoux M (2012) Disentangling the roles of history and local selection in shaping clinal variation of allele frequencies and gene expression in Norway spruce (Picea abies). Genetics 191:865–881
Chybicki IJ, Trojankiewicz M, Oleksa A, Dzialuk A, Burczyk J (2009) Isolation-by-distance within naturally established populations of European beech (Fagus sylvatica). Botany 87:791–798
Close TJ (1996) Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant 97:795–803
Comps B, Gömöry D, Letouzey J, Thiébaut B, Petit RJ (2001) Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157:389–397
Coop G, Witonsky D, Di Rienzo A, Pritchard JK (2010) Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–1423
de la Mata R, Zas R (2010) Transferring Atlantic maritime pine improved material to a region with marked Mediterranean influence in inland NW Spain: a likelihood-based approach on spatially adjusted field data. Eur J For Res 129:645–658
Derory J, Leger P, Garcia V, Schaeffer J, Hauser MT, Salin F, Luschnig C, Plomion C, Glossl J, Kremer A (2006) Transcriptome analysis of bud burst in sessile oak (Quercus petraea). New Phytol 170:723–738
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res 4:359–361
Eckert AJ, Bower AD, González-Martínez SC, Wegrzyn JL, Coop G, Neale DB (2010) Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae). Mol Ecol 19:3789–3805
European Communities (1999) Council directive 1999/105/EC of 22 December 1999 on the marketing of forest reproductive material. Off J Eur Commun 15. 1. 2000 17–L 11/40
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Gálvez L, González EM, Arrese-Igor C (2005) Evidence for carbon flux shortage and strong carbon/nitrogen interactions in pea nodules at early stages of water stress. J Exp Bot 56:2551–2561
Geburek T (2000) Effects of environmental pollution on the genetics of forest trees. In: Young A, Boshier D, Boyle T (eds) Forest conservation genetics, principles and practice. CSIRO Publishing, Colingwood and CABI Publishing, Oxon, pp 135–158
Gömöry D, Paule L (2011) Trade-off between height growth and spring flushing in common beech (Fagus sylvatica L.). Ann For Sci 68:975–984
Gömöry D, Paule L, Vyšný J (2007) Patterns of allozyme variation in western-Eurasian beeches. Bot J Linn Soc 154:165–174
Gömöry D, Ditmarová L, Hrivnák M, Jamnická G, Kmeť J, Krajmerová D, Kurjak D (2015) Differentiation in phenological and physiological traits in European beech (Fagus sylvatica L.). Eur J For Res 134:1075–1085
González-Martínez SC, Krutovsky KV, Neale DB (2006) Forest-tree population genomics and adaptive evolution. New Phytol 170:227–238
Grattapaglia D, Plomion C, Kirst M, Sederoff RR (2009) Genomics of growth traits in forest trees. Curr Opin Plant Biol 12:148–156
Grivet D, Sebastiani F, Alia R, Bataillon T, Torre S, Zabal-Aguirre M, Vendramin GG, González-Martínez SC (2011) Molecular footprints of local adaptation in two Mediterranean conifers. Mol Biol Evol 28:101–116
Hanewinkel M, Cullmann DA, Schelhaas M-J, Nabuurs G-J, Zimmermann NE (2013) Climate change may cause severe loss in the economic value of European forest land. Nat Clim Chang 3:203–207
Hemery GE (2008) Forest management and silvicultural responses to projected climate change impacts on European broadleaved trees and forests. Int For Rev 10:591–607
Jiménez JA, Alonso-Ramírez A, Nicolás C (2008) Two cDNA clones (FsDhn1 and FsClo1) up-regulated by ABA are involved indrought responses in Fagus sylvatica L. seeds. J Plant Physiol 165:1798–1807
Joost S, Bonin A, Bruford MW, Després L, Conord C, Erhardt G, Taberlet P (2007) A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol 16:3955–3969
Kawecki TJ, Ebert D (2004) Conceptual issues in local adaptation. Ecol Lett 7:225–1241
Kosová K, Vítámvás P, Prášil IT (2007) The role of dehydrins in plant response to cold. Biol Plant 51:601–617
Lalagüe H, Csilléry K, Oddou-Muratorio S, Safrana J, de Quattro C, Fady B, González-Martínez SC, Vendramin GG (2014) Nucleotide diversity and linkage disequilibrium at 58 stress response and phenology candidate genes in a European beech (Fagus sylvatica L.) population from southeastern France. Tree Genet Genomes 10:15–26
Lefèvre S, Wagner S, Petit RJ, de Lafontaine G (2012) Multiplexed microsatellite markers for genetic studies of beech. Mol Ecol Res 12:484–491
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst 27:237–277
Lotterhos KE, Whitlock MC (2014) Evaluation of demographic history and neutral parameterization on the performance of F ST outlier tests. Mol Ecol 23:2178–2192
Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Latalowa M, Litt T, Paule L, Roure JM, Tantau I, van der Knaap WO, Petit RJ, de Beaulieu J-L (2006) A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytol 171:199–222
Mátyás C (1996) Climatic adaptation of trees: rediscovering provenance tests. Euphytica 92:45–54
Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51:659–668
Merilä J, Crnokrak P (2001) Comparison of genetic differentiation at marker loci and quantitative traits. J Evol Biol 14:892–903
Mimura M, Aitken SN (2010) Local adaptation at the range peripheries of Sitka spruce. J Evol Biol 23:249–258
Mosca E, González-Martínez SC, Neale DB (2014) Environmental versus geographical determinants of genetic structure in two subalpine conifers. New Phytol 201:180–192
Müller M, Seifert S, Finkeldey R (2015) A candidate gene-based association study reveals SNPs significantly associated with bud burst in European beech (Fagus sylvatica L.). Tree Genet Genomes 11:116
Neale DB, Ingvarsson PK (2008) Population, quantitative and comparative genomics of adaptation in forest trees. Curr Opin Plant Biol 11:149–155
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122
Pastorelli R, Smulders MJM, Van’t Westende WPC, Vosman B, Giannini R, Vettori C, Vendramin GG (2003) Characterization of microsatellite markers in Fagus sylvatica L. and Fagus orientalis Lipsky. Mol Ecol Notes 3:76–78
Plomion C, Bastien C, Bogeat-Triboulot MB, Bouffier L, Déjardin A, Duplessis S, Fady B, Heuertz M, Le Gac AL, Le Provost G, Legue V, Lelu-Walter MA, Leplé JC, Maury S, Morel A, Oddou-Muratorio S, Pilate G, Sanchez L, Scotti I, Scotti-Saintagne C, Segura V, Trontin JF, Vacher C (2016) Forest tree genomics: 10 achievements from the past 10 years and future prospects. Ann For Sci 73:77–103
Pritchard JK, Stephens M, Donnelly P (2000a) Inference of population structure from multilocus genotype data. Genetics 155:945–959
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000b) Association mapping in structured populations. Am J Hum Genet 67:170–181
Rehfeldt GE, Ying CC, Spittlehouse DL, Hamilton DA (1999) Genetic responses to climate in Pinus concorta: niche breadth, climate change, and reforestation. Ecol Monogr 69:375–407
Rellstab C, Zoller S, Walthert L, Lesur I, Pluess AR, Graf R, Bodénès C, Sperisen C, Kremer A, Gugerli F (2016) Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Mol Ecol 25:5907–5924
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:103–106
Savolainen O, Pyhajärvi T, Knurr T (2007) Gene flow and local adaptation in trees. Ann Rev Ecol Evol Syst 38:595–619
Saxe H, Cannell MGR, Johnsen B, Ryan MG, Vourlitis G (2001) Tree and forest functioning in response to global warming. New Phytol 149:369–399
Scalfi M, Mosca E, Di Pierro EA, Troggio M, Vendramin GG, Sperisen C, La Porta N, Neale DB (2014) Micro- and macro-geographic scale effect on the molecular imprint of selection and adaptation in Norway spruce. PLoS ONE 9:e115499
Seifert S, Vornam B, Finkeldey R (2012) DNA sequence variation and development of SNP markers in beech (Fagus sylvatica L.). Eur J For Res 131:1761–1770
Shirakawa M, Ueda H, Koumoto Y, Fuji K, Nishiyama C, Kohchi T, Hara-Nishimura I, Shimada T (2014) Continuous vascular ring (COV1) is a trans-Golgi network-localized membrane protein required for Golgi morphology and vacuolar protein sorting. Plant Cell Physiol 55:764–772
Stephens M, Donelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169
Tanaka K, Tsumura Y, Nakamura T (1999) Development and polymorphism of microsatellite markers for Fagus crenata and the closely related species, F. japonica. Theor Appl Genet 99:11–15
Vitasse Y, Delzon S, Bresson CC, Michalet R, Kremer A (2009) Altitudinal differentiation in growth and phenology among populations of temperate-zone tree species growing in a common garden. Can J For Res 39:1259–1269
von Wuehlisch G, Krusche D, Muhs HJ (1995) Variation in temperature sum requirement for flushing of beech provenances. Silvae Genet 44:343–347
Vornam B, Gailing O, Derory J, Plomion C, Kremer A, Finkeldey R (2011) Characterisation and natural variation of a dehydrin gene in Quercus petraea (Matt.) Liebl. Plant Biol 13:881–887
Wachowiak W, Balk PA, Savolainen O (2009) Search for nucleotide diversity patterns of local adaptation in dehydrins and other cold-related candidate genes in Scots pine (Pinus sylvestris L.). Tree Genet Genomes 5:117–132
Welling A, Rinne P, Viherä-Aarnio A, Kontunen-Soppela S, Heino P, Palva ET (2004) Photoperiod and temperature differentially regulate the expression of two dehydrin genes during overwintering of birch (Betula pubescens Ehrh.). J Exp Bot 55:507–516
Williams MI, Dumroese RK (2013) Preparing for climate change: forestry and assisted migration. J For 111:287–297
Acknowledgements
The provenance experiment has been established through the realization of the project European Network for the Evaluation of the Genetic Resources of Beech for Appropriate Use in Sustainable Forestry Management (AIR3-CT94-2091) under the coordination of H.-J. Muhs and G. von Wühlisch. The experimental plot Tále was established by L. Paule. The study was supported by research grants of the Slovak Research and Development Agency APVV-0135-12 (DKr, MH, ĽD, JK, DKu, DG), APVV-0436-10 (ĽD, GJ), and Slovak Grant Agency for Science VEGA-2/0034/14 (ĽD, GJ). Technical assistance of G. Baloghová is greatly appreciated. We also thank to K. Willingham for linguistic correction.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krajmerová, D., Hrivnák, M., Ditmarová, Ľ. et al. Nucleotide polymorphisms associated with climate, phenology and physiological traits in European beech (Fagus sylvatica L.). New Forests 48, 463–477 (2017). https://doi.org/10.1007/s11056-017-9573-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11056-017-9573-9