Abstract
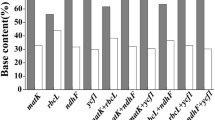
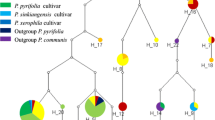
The study was conducted to evaluate the discriminatory potential of selected chloroplast-based DNA barcode regions for identifying and resolving phylogeny of the Indian bamboos. Among 11 chloroplast markers screened, only four, namely matK, rbcL, psbK-I and rps16-trnQ showed successful amplification in 88 genotypes of 30 Indian bamboo taxa under Bambuseae and Arundinarieae tribes. A total of 244 sequences were generated for the four chloroplast regions. Tree-based analysis demonstrated that none of the tested regions successfully discriminated the taxa under Bambuseae tribe. Importantly, our highly concerned Himalayan temperate bamboo species under Arundinarieae tribe, were successfully discriminated by matK locus with high bootstrap support (>60%). Sequence comparisons revealed that the discriminatory power demonstrated by matK region actually lies in the few unique fixed nucleotides (UFNs) despite the overall DNA polymorphism. Although, rps16-trnQ region was found to be the most polymorphic and revealed high genetic divergence among different taxonomic levels, it could not successfully discriminated the taxa with strong statistical support. In a taxonomically difficult plant group like bamboos, whose genome is relatively more complex and has a slow rate of molecular evolution, it is difficult to get a universal marker. Further, highly variable barcode regions utilized in other species may not be informative, and thus, the development of DNA barcodes for different taxonomic levels, such as lineages or tribes could be a viable approach.




Similar content being viewed by others
Availability of data and material
The sequence data is available in NCBI GenBank
References
Attigala L, Triplett JK, Kathriarachchi HS, Clark LG (2014) A new genus and a major temperate bamboo lineage of the Arundinarieae (Poaceae: Bambusoideae) from Sri Lanka based on a multi-locus plastid phylogeny. Phytotaxa 174(4):187. https://doi.org/10.11646/phytotaxa.174.4.1
Bi Y, Zhang M, Xue J, Dong R, Du Y, Zhang X (2018) Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci Rep. https://doi.org/10.1038/s41598-018-19591-9
Brandis D (1899) Biological notes on Indian Bamboos. Indian Forester 25(1):1–25
Cai ZM, Zhang YX, Zhang LN, Gao LM, Li DZ (2012) Testing four candidate barcoding markers in temperate woody bamboos (Poaceae: Bambusoideae). J Syst Evol 50:527–539. https://doi.org/10.1111/j.1759-6831.2012.00216.x
Campbell JNN (1985) Bamboo flowering patterns: a global view with special reference to East-Asia. Am Bamboo Soc J 6:17–35
Canavan S, Richardson DM, Visser V, Le Roux JJ, Vorontsova MS, Wilson JRU (2017) The global distribution of bamboos: assessing correlates of introduction and invasion. AoB Plants 9:plw078. https://doi.org/10.1093/aobpla/plw078
CBOL (2009) A DNA barcode for land plants. PNAS 106:12794–12797. https://doi.org/10.1073/pnas.0905845106
Chase MW, Fay MF (2009) Barcoding of plants and fungi. Science 325:682–683. https://doi.org/10.1126/science.1176906
Chase MW, Cowan RS, Hollingsworth PM, van den Berg C et al (2007) A proposal for a standardized protocol to barcode all land plants. Taxon 56:295–299. https://doi.org/10.1002/tax.562004
Chen S, Yao H, Han J et al (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5:e8613. https://doi.org/10.1371/journal.pone.0008613
Clark LG, Dransfield S, Triplett JK, Sanchez-Ken JG (2007) Phylogenetic relationships among the one-flowered, determinate genera of Bambuseae (Poaceae: Bambusoideae). Aliso 23(1):315–332. https://doi.org/10.5642/aliso.20072301.26
Cuenoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002) Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot 89:132–144. https://doi.org/10.3732/ajb.89.1.132
Das M, Bhattacharya S, Basak J, Pal A (2007) Phylogenetic relationships among the bamboo species as revealed by morphological characters and polymorphism analyses. Biol Plant 51:667–672. https://doi.org/10.1007/s10535-007-0140-7
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131. https://doi.org/10.1111/j.1365-294x.1995.tb00201.x
DeSalle R, Egan MG, Siddall M (2005) The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos Trans R Soc Lond B Biol Sci 360:1905–1916. https://doi.org/10.1098/rstb.2005.1722
Dev SA, Sijimol K, Prathibha PS, Sreekumar VB, Muralidharan EM (2020) DNA barcoding as a valuable molecular tool for the certification of planting materials in bamboo. 3 Biotech 10(2):59. https://doi.org/10.1007/s13205-019-2018-8
Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Cheng T, Guo J, Zhou S (2015) ycf1, the most promising plastid DNA barcode of land plants. Sci Rep 5:8348. https://doi.org/10.1038/srep08348
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull 19:11–15
Fay MF, Swensen SM, Chase MW (1997) Taxonomic Affinities of Medusagyne oppositifolia (Medusagynaceae). Kew Bull 52(1):111. https://doi.org/10.2307/4117844
Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW et al (2008) Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3(7):2802. https://doi.org/10.1371/journal.pone.0002802
Ford CS, Ayres KL, Toomey N, Haider N et al (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc 159:1–11. https://doi.org/10.1111/j.1095-8339.2008.00938.x
Gaut BS, Clark LG, Wendel JF, Muse SV (1997) Comparisons of the molecular evolutionary process at rbcL and ndhF in the grass family (Poaceae). Mol Biol Evol 14(7):769–777. https://doi.org/10.1093/oxfordjournals.molbev.a025817
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysisprogram for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. https://doi.org/10.14601/Phytopathol_Mediterr-14998u1.29
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc B 270:313–321. https://doi.org/10.1098/rspb.2002.2218
Hollingsworth ML, Clark A, Forrest LL, Richardson J et al (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour 9:439–457. https://doi.org/10.1111/j.1755-0998.2008.02439.x
Hosein FN, Austin N, Maharaj S, Johnson W, Rostant L, Ramdass AC, Rampersad SN (2017) Utility of DNA barcoding to identify rare endemic vascular plant species in Trinidad. Ecol Evol 7(18):7311–7333. https://doi.org/10.1002/ece3.3220
Huelsenbeck JP, Ronquist F (2001) MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755. https://doi.org/10.1093/bioinformatics/17.8.754
ISFR (2019) India State of Forest Report, vol 1, 16th edn. Forest Survey of India, Ministry of Environment Forests and Climate Change, Dehradun, India
Janzen DH (1976) Why bamboos wait so long to flower. Annu Rev Ecol Evol S 7:347–391. https://doi.org/10.1146/annurev.es.07.110176.002023
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Krawczyk K, Nobis M, Myszczynski K, Klichowska E, Sawicki J (2018) Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci Rep 8:1924. https://doi.org/10.1038/s41598-018-20399-w
Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2(6):508. https://doi.org/10.1371/journal.pone.0000508
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. PNAS 102:8369–8374. https://doi.org/10.1073/pnas.0503123102
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Lahaye R, Van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. PNAS 105(8):2923–2928. https://doi.org/10.1073/pnas.0709936105
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47(1):256–259. https://doi.org/10.1093/nar/gkz239
Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S (2014) Plant DNA barcoding: from gene to genome. Biol Rev 90:157–166. https://doi.org/10.1111/brv.12104
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Makita A (1992) Survivorship of a monocarpic bamboo grass, Sasa kurilensis, during the early regeneration process after mass flowering. Ecol Res 7:245–254. https://doi.org/10.1007/BF02347093
Meena RK, Bhandhari MS, Barhwal S, Ginwal HS (2019) Genetic diversity and structure of Dendrocalamus hamiltonii natural metapopulation: a commercially important bamboo species of northeast Himalayas. 3 Biotech 9:60. https://doi.org/10.1007/s13205-019-1591-1
Ohrnberger D, Georrings J (1986) The bamboos of the World. International Book Publishers, Dehra Dun
Olmstead RG, Sweere JA (1994) Combining data in phylogenetic systematics: an empirical approach using three molecular data sets in the Solanaceae. Syst Biol 43:467–481. https://doi.org/10.2307/2413546
Parks M, Cronn R, Liston A (2009) Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol 7:84–100. https://doi.org/10.1186/1741-7007-7-84
Peterson PM, Romaschenko K, Soreng RJ (2014) A laboratory guide for generating DNA barcodes in grasses: a case study of Leptochloa s.l. (Poaceae: Chloridoideae). Webbia 69(1):1–12. https://doi.org/10.1186/1741-7007-7-84
Pettengill JB, Neel MC (2010) An evaluation of candidate plant DNA barcodes and assignment methods in diagnosing 29 species in the genus Agalinis (Orobanchaceae). Am J Bot 97:1391–1406. https://doi.org/10.3732/ajb.0900176
Sang T, Crawford D, Stuessy T (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot 84(8):1120. https://doi.org/10.2307/2446155
Sarma KK, Pathak KC (2004) Leaf and culm sheath morphology of some important bamboo species of Assam. J Bamboo Rattan 3(3):265–281. https://doi.org/10.1163/156915904176525
Smith SA, Donoghue MJ (2008) Rates of molecular evolution are linked to life history in flowering plants. Science 322(5898):86–89. https://www.jstor.org/stable/20144950
Stapleton CMA (2009) Molecular phylogeny of Asian woody bamboos: review for the Flora of China. Bamboo Sci Cult J Amer Bamboo Soc 22(1):5–25
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109. https://doi.org/10.1007/BF00037152
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Tate JA, Simpson BB (2003) Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot 28:723–737. https://doi.org/10.1043/02-64.1
Triplett JK, Clark LG (2010) Phylogeny of the temperate woody bamboos (Poaceae: Bambusoideae) with an emphasis on Arundinaria and allies. Syst Bot 35:102–120. https://doi.org/10.1600/036364410790862678
Woudenberg JHC, Seidl MF, Groenewald JZ, de Vries M, Stielow JB, Thomma BPHJ, Crous PW (2015) Alternaria section Alternaria: species, formae speciales or pathotypes? Stud Mycol 82:1–21. https://doi.org/10.1016/j.simyco.2015.07.001
Wu FH, Kan DP, Lee SB, Daniell H et al (2009) Complete nucleotide sequence of Dendrocalamus latiflorus and Bambusa oldhamii chloroplast genomes. Tree Physiol 29(6):847–856. https://doi.org/10.1093/treephys/tpp015
Rozas J (2009) DNA Sequence polymorphism analysis using DnaSP. In: Posada D (ed) Bioinformatics for DNA sequence analysis. Methods in molecular biology series Vol. 537. Humana Press, pp 337–350. https://doi.org/10.1007/978-1-59745-251-9_17
Yang JB, Tang M, Li HT, Zhang ZR, Li DZ (2013) Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol 13(1):84. https://doi.org/10.1186/1471-2148-13-84
Yamane K, Kawahara T (2005) Intra- and inter-specific phylogenetic relationships among diploid Triticum-Aegilops species (Poaceae) based on base-pair substitutions, indels, and microsatellites in chloroplast noncoding sequences. Am J Bot 92:1887–1898. https://doi.org/10.3732/ajb.92.11.1887
Zeng CX, Zhang YX, Triplett JK, Yang JB, Li DZ (2010) Large multi-locus plastid phylogeny of the tribe Arundinarieae (Poaceae: Bambusoideae) reveals ten major lineages and low rate of molecular divergence. Mol Phylogenet Evol 56:821–839. https://doi.org/10.1016/j.ympev.2010.03.041
Zhang YJ, Ma PF, Li DZ (2011) High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae). PLoS ONE 6(5):e20596. https://doi.org/10.1371/journal.pone.0020596
Acknowledgements
The work was financially supported by Indian Council of Forestry Research and Education (ICFRE), Dehradun as a research project [OG-49/CR-19]. We thank to the Director Forest Research Institute, Dehradun for providing facility for laboratory and field works. We are also very grateful to the Directors of different institute, namely State Forest Research Institute (SFRI) Itanagar, Rain Forest Research Institute (RFRI) Jorhat, Forest Research Centre for Bamboo and Rattan (FRC-BR), Aizawl (Mizoram) for providing access to their bambusetum. The forest departments of Uttarakhand and north eastern states of India are also duly acknowledged for their assistance and permissions in surveys and sample collection from the forest area under their jurisdiction. Finally, we are thankful to Dr. H. B. Naithani (a retired bamboo taxonomist) for his contribution in correct taxonomic identification of the bamboo species.
Funding
The work was financially supported by Indian Council of Forestry Research and Education (ICFRE) as research project [OG-49/CR-19].
Author information
Authors and Affiliations
Contributions
RKM, MSB, and RS involved in project conceptualization, execution, sample collection, and manuscript writing; NN and NU conducted the laboratory work; RKN involved in taxonomic verification of the sample specimen; RKM, AS and SP carried out sequence data analysis; and HSG was the mentor and involved in manuscript writing and editing.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical approval
All necessary permissions were obtained for sample collection from concerned state forest department.
Consent to participate
All authors agreed to participate for submission of the research to the journal.
Consent for publication
All the authors have approved the manuscript for submission and publication in journal.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meena, R.K., Negi, N., Uniyal, N. et al. Chloroplast-based DNA barcode analysis indicates high discriminatory potential of matK locus in Himalayan temperate bamboos. 3 Biotech 10, 534 (2020). https://doi.org/10.1007/s13205-020-02508-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02508-7