Abstract
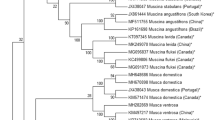
The Muscid flies belonging to the order Diptera, Family Muscidae, are notoriously famous for their ability to transmit disease-causing organisms to humans, livestock, and even plants. The huge species count of this group not only makes the classical method of identification laborious but also error-prone. Therefore, in this study, we analysed cytochrome oxidase 1 (cox1) barcodes to test their ability to discriminate between studied taxonomic groups. In our study, we used a total of 42 specimens, excluding the outgroup, of 14 species belonging to 5 genera representing 4 subfamilies were used. 11 specimens were collected from the lower Gangetic Plains of West Bengal, India, including the vulnerable Sagar Islands, each showing a very high AT content (average = 69%), especially in the 1st codon position. Cox1 barcodes were able to distinguish between species with very high interspecific genetic divergence. The percentage of intraspecific genetic divergences ranges from 0% to 2.2% for the studied dataset, with the interspecific range being very high, from 0.58% to 28.8%. 1000 bootstrapped Maximum likelihood (ML) with Kimura-2-parameter phylogenetic tree and similar bootstrap-based IQ-Tree was constructed based on cox1 gene, for clarity. The ML tree is used to understand the delimitation of the species using Poisson Tree Process (PTP) analysis. The phylogenetic and delimitation study determined that most of the genus level clusters are monophyletic. The subfamily Muscinae showed non-monophyletic lineage as seen by the separation of genera Musca from genera Haematobia in both ML and IQ-Tree.



Similar content being viewed by others
Change history
16 December 2022
The original online version of this article was revised to correct the ascension number of some raw data from OL444726 to OL539726.
15 December 2022
A Correction to this paper has been published: https://doi.org/10.1007/s42690-022-00934-z
References
Banerjee D, Kumar V, Maity A et al (2015) Identification through DNA barcoding of Tabanidae (Diptera) vectors of surra disease in India. Acta Trop 150:52–58
Buckman RS, Mound LA, Whiting MF (2013) Phylogeny of thrips (Insecta: Thysanoptera) based on five molecular loci. Syst Entomol 38:123–133
Chakraborty R, Singha D, Kumar V et al (2019) DNA barcoding of selected Scirtothrips species (Thysanoptera) from India. Mitochondrial DNA Part B 4:2710–2714
Changbunjong T, Weluwanarak T, Samung Y, Ruangsittichai J (2016) Molecular identification and genetic variation of hematophagous flies, (Diptera: Muscidae: Stomoxyinae) in Thailand based on cox1 barcodes. J Asia-Pac Entomol 19:1117–1123. https://doi.org/10.1016/j.aspen.2016.10.006
Cywinska A, Hunter FF, Hebert PD (2006) Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol 20:413–424
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772–772
de Carvalho CJB, de Mello-Patiu CA (2008) Key to the adults of the most common forensic species of Diptera in South America. Rev Bras Entomol 52:390–406
DeSalle R, Egan MG, Siddall M (2005) The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos Trans R Soc B Biol Sci 360:1905–1916. https://doi.org/10.1098/rstb.2005.1722
Evenhuis NL, Pape T (2022) Systema Dipterorum, Version 3.8, 2022. Version 3.8
Folmer O, Black M, Hoeh W et al (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Grzywacz A, Hall MJ, Pape T, Szpila K (2017) Muscidae (Diptera) of forensic importance—an identification key to third instar larvae of the western Palaearctic region and a catalogue of the muscid carrion community. Int J Legal Med 131:855–866
Grzywacz A, Trzeciak P, Wiegmann BM et al (2021) Towards a new classification of Muscidae (Diptera): a comparison of hypotheses based on multiple molecular phylogenetic approaches. Syst Entomol 46:508–525
Haseyama KL, Wiegmann BM, Almeida EA, de Carvalho CJ (2015) Say goodbye to tribes in the new house fly classification: a new molecular phylogenetic analysis and an updated biogeographical narrative for the Muscidae (Diptera). Mol Phylogenet Evol 89:1–12
Hebert PD, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci 270:313–321
Iwasa M, Ishiguro N (2010) Genetic and morphological differences of Haematobia irritans and H. exigua, and molecular phylogeny of Japanese Stomoxyini flies (Diptera, Muscidae). Med Entomol Zool 61:335–344. https://doi.org/10.7601/mez.61.335
Johnson M, Zaretskaya I, Raytselis Y et al (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9
Kettle DS (1995) Muscidae and Fanniidae. Med Vet Entomol 248–267
Kim YH, Shin SE, Ham CS et al (2014) Molecular identification of necrophagous muscidae and sarcophagidae fly species collected in Korea by mitochondrial cytochrome c oxidase subunit i nucleotide sequences. Sci World J. https://doi.org/10.1155/2014/275085
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547
Kutty SN, Pape T, Pont A et al (2008) The Muscoidea (Diptera: Calyptratae) are paraphyletic: Evidence from four mitochondrial and four nuclear genes. Mol Phylogenet Evol 49:639–652. https://doi.org/10.1016/j.ympev.2008.08.012
Kutty SN, Pont AC, Meier R, Pape T (2014) Complete tribal sampling reveals basal split in Muscidae (Diptera), confirms saprophagy as ancestral feeding mode, and reveals an evolutionary correlation between instar numbers and carnivory. Mol Phylogenet Evol 78:349–364
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Low VL, Tan TK, Prakash BK et al (2017) Contrasting evolutionary patterns between two haplogroups of Haematobia exigua (Diptera: Muscidae) from the mainland and islands of Southeast Asia. Sci Rep 7:1–9
Lunt DH, Hyman BC (1997) Animal mitochondrial DNA recombination. Nature 387:247–247
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 gateway computing environments workshop (GCE). Ieee, pp 1–8
Minh BQ, Schmidt HA, Chernomor O et al (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534
Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Pape T, Blagoderov V, Mostovski MB (2011) Order Diptera Linnaeus, 1758. In: Zhang, Z.-Q.(Ed.) Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness. Zootaxa 3148:222–229
Rivera J, Currie DC (2009) Identification of Nearctic black flies using DNA barcodes (Diptera: Simuliidae). Mol Ecol Resour 9(Suppl s1):224–236. https://doi.org/10.1111/j.1755-0998.2009.02648.x
Schühli G, De Carvalho C, Wiegmann B (2007) Molecular phylogenetics of the Muscidae (Diptera:Calyptratae): New ideas in a congruence context. Invertebr Syst. https://doi.org/10.1071/IS06026
Shi Y-W, Liu X-S, Wang H-Y, Zhang R-J (2009) Seasonality of insect succession on exposed rabbit carrion in Guangzhou, China. Insect Sci 16:425–439
Skidmore P (1985) The biology of the Muscidae of the world. Springer Science & Business Media
Srivathsan A, Meier R (2012) On the inappropriate use of Kimura-2-parameter (K2P) divergences in the DNA-barcoding literature. Cladistics 28:190–194. https://doi.org/10.1111/j.1096-0031.2011.00370.x
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci 101:11030–11035
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Taylor DB, Moon RD, Mark DR (2012) Economic impact of stable flies (Diptera: Muscidae) on dairy and beef cattle production. J Med Entomol 49:198–209
Tyagi K, Kumar V, Singha D et al (2017) DNA Barcoding studies on Thrips in India: Cryptic species and Species complexes. Sci Rep 7:1–14
Wells JD, Sperling FA (2001) DNA-based identification of forensically important Chrysomyinae (Diptera: Calliphoridae). Forensic Sci Int 120:110–115. https://doi.org/10.1016/s0379-0738(01)00414-5
Xiong F, Guo Y, Luo B et al (2012) Identification of the forensically important flies (Diptera: Muscidae) based on cytochrome oxidase subunit I (COI) gene in China. Afr J Biotechnol 11:10912–10918. https://doi.org/10.4314/ajb.v11i48
Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876
Zimin LS, Elberg KY (1988) Family Muscidae. Keys Insects Eur Part USSR 5:839–974
Acknowledgements
The authors are thankful to The Director, Zoological Survey of India, for her moral support and giving provisions for necessary facilities. Authors are also grateful to Dr. Vikas Kumar, Scientist – E, Molecular Systematics Division, Zoological Survey of India, for giving the facilities to work on the molecular systematics portion of this paper.
Funding
The work is funded partially by The Zoological Society, Kolkata (grant no: ZSK/NIA-B1/ 2021) to the first author. The core financing facilities from the Zoological Survey of India is responsible for supplying the laboratory and chemical facilities.
Author information
Authors and Affiliations
Contributions
Sample collection and preservation: KM, DP; Methodology and data accumulation: DG, OK, AM; Data analysis and manuscript writing: DG, OK, Manuscript review: AN, OK; GIS map generation: SS; Laboratory material and equipment provided by: DB.
Corresponding author
Ethics declarations
Conflict of Interest
The authors here show no conflict of interest in the work done.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghosh, D., Kar, O., Pramanik, D. et al. Molecular identification and characterization of Muscid flies (Diptera: Muscidae) of medico-veterinary importance from the Gangetic plains of Eastern India. Int J Trop Insect Sci 42, 3759–3769 (2022). https://doi.org/10.1007/s42690-022-00900-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42690-022-00900-9