Phylogenomics and the first higher taxonomy of Placozoa, an ancient and enigmatic animal phylum
- 1Department of Biology, St. Francis College, Brooklyn, NY, United States
- 2Sackler Institute for Comparative Genomics, American Museum of Natural History, New York, NY, United States
- 3Division of Invertebrate Zoology, American Museum of Natural History, New York, NY, United States
- 4Center for Genomics and Systems Biology, New York University, New York, NY, United States
- 5Richard Gilder Graduate School, American Museum of Natural History, New York, NY, United States
- 6Institute of Animal Ecology, Stiftung Tierärztliche Hochschule Hannover, Hannover, Germany
- 7Bigelow Laboratory for Ocean Sciences, Boothbay, ME, United States
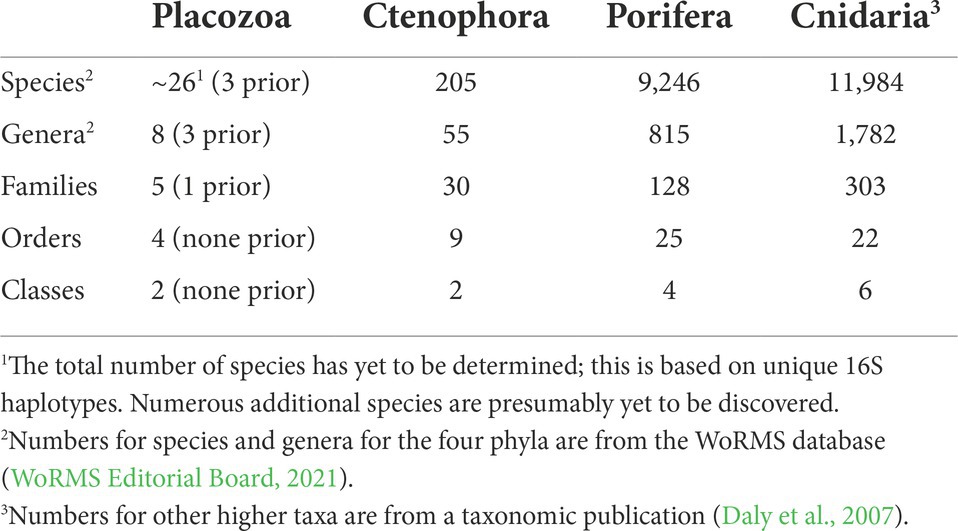
Placozoa is an ancient phylum of extraordinarily unusual animals: miniscule, ameboid creatures that lack most fundamental animal features. Despite high genetic diversity, only recently have the second and third species been named. While prior genomic studies suffer from incomplete placozoan taxon sampling, we more than double the count with protein sequences from seven key genomes and produce the first nuclear phylogenomic reconstruction of all major placozoan lineages. This leads us to the first complete Linnaean taxonomic classification of Placozoa, over a century after its discovery: This may be the only time in the 21st century when an entire higher taxonomy for a whole animal phylum is formalized. Our classification establishes 2 new classes, 4 new orders, 3 new families, 1 new genus, and 1 new species, namely classes Polyplacotomia and Uniplacotomia; orders Polyplacotomea, Trichoplacea, Cladhexea, and Hoilungea; families Polyplacotomidae, Cladtertiidae, and Hoilungidae; and genus Cladtertia with species Cladtertia collaboinventa, nov. Our likelihood and gene content tree topologies refine the relationships determined in previous studies. Adding morphological data into our phylogenomic matrices suggests sponges (Porifera) as the sister to other animals, indicating that modest data addition shifts this node away from comb jellies (Ctenophora). Furthermore, by adding the first genomic protein data of the exceptionally distinct and branching Polyplacotoma mediterranea, we solidify its position as sister to all other placozoans; a divergence we estimate to be over 400 million years old. Yet even this deep split sits on a long branch to other animals, suggesting a bottleneck event followed by diversification. Ancestral state reconstructions indicate large shifts in gene content within Placozoa, with Hoilungia hongkongensis and its closest relatives having the most unique genetics.
Introduction
Despite its ameboid gestalt and utter lack of typical animal features, Placozoa is indeed a phylum of true metazoan animals (Schierwater, 2005). Because of its extremely simple bauplan and morphological uniformity, only one accepted species was described for over a century (Schulze, 1883; Grell, 1972; Eitel et al., 2018; Osigus et al., 2019). This lack of easy-to-observe morphological variation did not help systematists divine almost any of the cryptic diversity harbored in this ancient group (Voigt et al., 2004; Miller and Ball, 2005; Pearse and Voigt, 2007; Eitel et al., 2013; Paknia and Schierwater, 2015; Schierwater et al., 2021a). The phylum has been monotypic until recently and is one of the oldest in the animal kingdom, which molecular data suggests to be well over 500 million years old (Hedges et al., 2004; Peterson and Butterfield, 2005) and composed of many highly diverged lineages (Voigt et al., 2004; Eitel et al., 2013). In the current phylogenomics era (DeSalle et al., 2020), the time has come to more confidently determine the relationships among placozoan lineages (Schierwater et al., 2021b). Here we substantially add to the amount of Placozoa nuclear genomic data, produce the first nuclear phylogenomic reconstruction of the major lineages, and establish higher Linnaean taxonomy within the phylum.
Historical background
In 1883 the first species — Trichoplax adhaerens — was described and suggested to be at the base of the animal tree (Schulze, 1883). After a decade, Treptoplax reptans was described (Monticelli, 1893, 1895). Unfortunately, the living T. reptans specimens from Monticelli (1893) died and have never been resampled in the field; the characteristics identified by Monticelli, however, have been doubted and this species is therefore currently unaccepted (Syed and Schierwater, 2002a; see also WoRMS Editorial Board, 2021), although it could be rare or extinct.
Placozoans were subsequently misclassified as adult acoel turbellarians (Noll, 1890; Syed and Schierwater, 2002a) and cnidarian larvae (Krumbach, 1907). Thus, decades passed without studies on Placozoa (Grell, 1971a; Birstein, 1989). In the 1960’s the lineage was rediscovered (Kuhl and Kuhl, 1966) and soon considered distinct again (Grell, 1971a,b, 1972; Grell and Benwitz, 1974). Significantly, the phylum Placozoa was formally erected (Grell, 1971a; Birstein, 1989). More modern morphological studies have identified ultrastructure characters that differ between some placozoans (Guidi et al., 2011; Romanova et al., 2021), but diagnostic morphological characters remain limited.
Placozoa molecular background and the sister to other Metazoa
In the early 2000s, molecular data began to show an unexpected amount of genetic diversity in this “phylum of one” (Voigt et al., 2004; Signorovitch et al., 2007; Eitel et al., 2013). Ongoing 16S analyses consistently revealed new placozoan haplotypes, with genetic distances on par with higher taxa in other animal phyla (Eitel and Schierwater, 2010). While this immense cryptic diversity has become more and more obvious in the last two decades, the scientific community has remained hesitant (generally for good reasons) to accept the formal description of animal species using only molecular markers (Voigt et al., 2004; Signorovitch et al., 2007; Eitel et al., 2013).
This does not mean that Placozoa researchers have been unable to communicate about these animals: a set of clade names has been regularly used since early ribosomal DNA studies to reference various lineages (Voigt et al., 2004). These clades have been examined several times with additional markers and mitogenomics (Signorovitch et al., 2007; Eitel et al., 2013; Simion et al., 2017; Whelan et al., 2017). Still, the stability and formality of incorporating Linnaean taxonomy alongside these molecularly-identified clades needs to continue to advance, as has recently occured for two new genera (Eitel et al., 2018; Osigus et al., 2019).
Most early phylogenetic and more modern phylogenomic studies focused on the position of Placozoa within animals, i.e., Metazoa (Peterson and Eernisse, 2001; Wallberg et al., 2004; Dellaporta et al., 2006; Telford, 2006; Erpenbeck et al., 2007; Signorovitch et al., 2007; Carr et al., 2008; Ryan et al., 2013; Chang et al., 2015; Simion et al., 2017; Whelan et al., 2017; Neumann et al., 2021a,b). The relationships between the principal animal lineages (Bilateria, Cnidaria, Ctenophora, Placozoa, and Porifera) have remained hotly debated since almost the beginning of evolutionary thinking (Schulze, 1883; Neumann et al., 2021a,b). Nearly all possible combinations have been found empirically, ultimately providing exceptionally short-lived hypotheses (Schierwater et al., 2009, 2010; Neumann et al., 2021b). Generally, either Porifera, Ctenophora, or Placozoa (Littlewood et al., 1998; some trees in Pett et al., 2019) is inferred as the sister of all other Metazoa (SOM; Dellaporta et al., 2006; Ryan et al., 2013; Chang et al., 2015; Simion et al., 2017; Whelan et al., 2017).
Phylogenomic reconstructions of early metazoans have placed Placozoa as sister to Cnidaria (Laumer et al., 2018), Bilateria+Cnidaria (Dunn and Ryan, 2015; Feuda et al., 2017; Simion et al., 2017), Bilateria+Coelenterata (i.e., Cnidaria+Ctenophora) (Philippe et al., 2009), and even to Porifera+Coelenterata, together forming the sister group to Bilateria (Schierwater et al., 2009). Ultimately, most phylogenomic studies suggest that Placozoa is probably not the SOM, though over 25% of our prior combined morphological and phylogenomic analyses do show Placozoa as the SOM (see Neumann et al., 2021a,b). If the majority of studies are correct, the morphological simplicity of Placozoa may be caused by major reductions of features, such as losing the basal membrane or other specialized cells, e.g., neurons and muscle cells (Nielsen, 2019). However, it has also been forcefully argued that placozoan morphology is not principally due to reductions in anatomical features: Instead, the most simple bauplan and one of the smallest nuclear genomes of all free-living animals have been interpreted as strong ancestral characters. Based on this, the “placula hypothesis” suggests the bauplans of Porifera, Cnidaria, and other animal phyla are derived from a common ancestor closely resembling Placozoa (Bütschli, 1884; Syed and Schierwater, 2002a,b; Schierwater et al., 2010). While T. adhaerens has one of the largest mitochondrial genomes of all animals (~43.000 bp), which has been seen as another argument for a Placozoa-sister hypothesis (Dellaporta et al., 2006), the recently discovered Polyplacotoma mediterranea (the sister to all other known placozoans) has a mitochondrial genome that is just over half that size (~23.000 bp), which is more typical for other Metazoa (Osigus et al., 2019).
All this poses the question of how to interpret morphology and molecules. Most authors disregard morphology when building trees regarding the SOM. However, we have shown that morphology can strongly impact phylogenomic results regarding the SOM (Neumann et al., 2021a,b). We are convinved that both data types contain true signal at this deep evolutionary scope, and that the answers about early animal evolution lie in a combination of data types. Accordingly, we include morphology in our phylogenomic matrix comparing metazoan phyla.
Current and future systematics of Placozoa
Placozoan systematics has great potential to advance, given that these animals are undersampled, yet have substantial genetic variability. For this advancement, new data are critical. With next-generation sequencing, molecular data appear to be the most abundant data source for placozoan systematics. Genomic data allow us to describe species, solidify relationships between major lineages, and establish higher-level classifications, as we do here.
While genomes may be the easiest route towards systematizing the phylum, getting even a few has taken massive effort. The first genome (T. adhaerens) was published in 2008 (Srivastava et al., 2008), with five others (Hoilungia hongkongensis, H2, H4, H6, and H11) sequenced a decade later (Eitel et al., 2018; Kamm et al., 2018; Laumer et al., 2018). The additional data published in our current study more than double the amount of data for phylogenomics.
It should be emphasized that collecting placozoans in the field is difficult (Pearse and Voigt, 2007). Culturing difficulties and placozoans’ miniscule size also make it challenging to obtain enough DNA or RNA for genome sequencing, because this usually requires pooling 1000’s of clonal replicates per placozoan lineage, as we did for most of the genomes presented here (Srivastava et al., 2008; Eitel et al., 2011, 2018). Recent work achieved partial transcriptomes and genomes using only a handful of individuals (Laumer et al., 2018). We take this a bit further, and base a placozoan genome sequence on a single unculturable individual (H24, see Table 1).
Excitingly, genome sequencing (Srivastava et al., 2008), years of collecting and culturing (Grell, 1971a; Schierwater et al., 2010; Eitel et al., 2013), anatomical ultrastructure analysis (Grell and Benwitz, 1971, 1981; Smith et al., 2014; Romanova et al., 2021), and phylogenetics (Voigt et al., 2004; Dellaporta et al., 2006; Signorovitch et al., 2007; Eitel et al., 2013) have made it possible to describe genetically isolated, yet morphologically indistinguishable doppelgängers as species. After almost 140 years of taxonomic vacuum, these breakthroughs gave us three described species of Placozoa: T. adhaerens (Schulze, 1883; Srivastava et al., 2008), H. hongkongensis (Eitel et al., 2018), and P. mediterranea (Osigus et al., 2019). This is surely only the beginning: most lineages remain undescribed (Eitel et al., 2013).
With new species being described, it is now time to confirm relationships within Placozoa in order to create a stable Linnaean taxonomy. We here present the first comprehensive phylogenomics for Placozoa, and create the first Linnaean taxonomy for this animal phylum, as well as describe a new species.
It is important to note that phylogenetics and phylogenomics have empowered major updates to the existing taxonomies of other phyla, such as Porifera and Cnidaria. These include the addition of the poriferan class Homoscleromorpha (Gazave et al., 2010), formally incorporating Myxozoa into Cnidaria (Jiménez-Guri et al., 2007; Nesnidal et al., 2013; Feng et al., 2014), and classifying Staurozoa as a new cnidarian class (Marques and Collins, 2005; Miranda et al., 2016). The phylogenetic relationships within Ctenophora will likely continue to evolve with future studies and increased taxon sampling (Podar et al., 2001; Simion et al., 2015; Whelan et al., 2017). There have also been rare exciting discoveries of new animal groups since the 1980s, such as the phyla Loricifera (Kristensen, 1983), Cycliophora (Funch and Kristensen, 1995), and Micrognathozoa, which is sometimes considered a phylum (Kristensen and Funch, 2000; Nielsen, 2012). Still, our current study presents the only complete higher taxonomy to be produced for an entire animal phylum in the 21st century.
Materials and methods
Nomenclature
For much of the manuscript, we reference the species names, Trichoplax adhaerens, Hoilungia hongkongensis, and Polyplacotoma mediterranea, as well as their genera and the family Trichoplacidae from prior research (Schulze, 1883; Eitel et al., 2018; Osigus et al., 2019), coupled with the informal clade names used for known 16S haplotypes (Eitel et al., 2013). However, later in this paper, we revise and expand on this taxonomy.
Specimens
See Table 1 for specimen information. Collecting and culturing followed prior protocols (Eitel and Schierwater, 2010).
Genome sequencing and assembly
We sequenced seven new Placozoa genomes. Additional Placozoa and outgroup proteomes were downloaded from publicly available datasets to produce our phylogenomic matrix (Table 1 and Supplementary Table 1). Of the newly sequenced genomes, we provide the phylogenomic matrix for all placozoans as amino acid data (Supplementary Data 1). The genomes and complete proteomes will be presented in more detail in Kamm et al. (in prep).
For the seven new genomes, DNA was extracted as described in Kamm et al. (2018). In the case of H24 and P. mediterranea, the genomic DNA had to be enriched prior to library preparation because of the lower quantity of available clonal animals. This has already been described for the mitochondrial genome assembly for P. mediterranea (Osigus et al., 2019) and for H24 (Miyazawa et al., 2021). Libraries were prepared at the New York Genome Center using an Illumina TruSeq PCR-free 450 bp procedure, then sequenced using an Illumina HiSeq2500, 2 × 125 bp.
Reads were filtered and trimmed in Trimmomatic (Bolger et al., 2014) and further corrected using the hammer module in SPAdes (Prjibelski et al., 2020). Next, the genomes were assembled with Platanus 1.2.4 (Kajitani et al., 2014) using a maximum distance for bubble crush (−u) of 0.15, and a maximum distance for branch cutting (−d) of 0.4. All other parameters to Platanus during assembly, scaffolding, and gap closing were defaults.
Genome annotation
Following genome assembly, filtering to remove duplicate contigs and endosymbiont sequences was applied as in prior work (Kamm et al., 2018). Our procedure added one additional pre-filtering step using MyCC version 2017 (Lin and Liao, 2016) to cluster contigs prior to identification and removal of outliers. Genome filtering involved removing all contigs shorter than 2kbp, using all-versus-all BLAST to identify redundant contigs following prior work (Kamm et al., 2018), running MyCC to cluster remaining contigs, identification of clusters of bacterial contigs by BLAST of outlier clusters against nr, and filtering the remaining contigs against a collection of sequences from the rickettsiales as in prior work (Kamm et al., 2018). Structural annotation and genome completeness measures were run on the filtered genomes.
Structural annotation of the new genomes reported here used Maker v2.31.10 (Campbell et al., 2014). RepeatModeler v1.0.11 (Smit and Hubley, 2015) was used to identify species-specific repeats, and those were used during the Maker repeat masking procedure. The Maker pipeline was run with the ab initio predictors Augustus and GeneMark that had been trained on placozoan genomes like described before (Kamm et al., 2018). Evidence given to Maker consisted of selected UniProt proteomes and placozoan transcriptome predictions as described in prior work (Kamm et al., 2018).
BUSCO
Genome gene coverage was assessed using BUSCO (Simão et al., 2015) with the metazoa_odb10 954 conserved single copy gene set (Table 1). The e-value cutoff was set to 1e-5, and the “--long” option added maximum precision. Genome statistics including BUSCO scores, N50, scaffold and exon information are summarized in Supplementary Table 2.
Orthology
Orthologs were defined with our prior phylogenomic pipeline (Lee et al., 2011) using OrthologID (Chiu et al., 2006). In short, input peptide sequences from all genomes were pairwise compared against each other using BLASTP (Altschul et al., 1990). The e-value scores were fed into the MCL clustering algorithm (Enright et al., 2002) to define gene family clusters. Clusters were aligned using MAFFT v7.407 (Katoh et al., 2002; Katoh and Standley, 2013). A parsimony tree was reconstructed for each set of alignments using TNT v1.1 (Goloboff et al., 2008). Orthologs were determined from identified duplication events in gene family trees (Lee et al., 2011). Ortholog alignments were then concatenated. We have summarized information on all 15,849 identified orthologs in a single file (Supplementary Data 2).
Phylogenomics and gene content
Two phylogenomic trees (including and excluding a morphological matrix), a gene content tree, and ancestral state reconstructions of gene content were conducted to assess placozoan evolution. Reconstructions and analyses were visualized using R version 4.0.0 (R Core Team, 2020) with ggtree (Yu et al., 2017) and ggplot2 (Wickham, 2009). Trees were based on the amino acid sequence matrix in Supplementary Data 1, and rooted using Capsaspora owczarzaki based on current understanding of our outgroup and ingroup relationships (Ruiz-Trillo et al., 2004; Sebé-Pedrós et al., 2013; Suga et al., 2013; Laumer et al., 2019; Fernández and Gabaldón, 2020; Pandey and Braun, 2020).
For our primary phylogenomic tree, we added a morphology matrix to the genomic data matrix for a combined analysis. We reconstructed the maximum likelihood topology for most major Placozoa lineages using C10 + GTR20 + F + G in IQTree 2.1.3 with ultrafast bootstrap approximation and the MK model used for the morphological data partition (Hoang et al., 2018; Minh et al., 2020). Note that C10 is the maximum likelihood equivalent of the Bayesian CAT models. Our matrix included 2,309,771 molecular characters spread across 1,882 orthologs. This was a reduction of our total dataset to mitigate the effects of missing data: we required ~90% of taxa (specifically > = 25 of the total 28 taxa) to contain an ortholog for inclusion in our final matrices. We ran this molecular matrix with and without 51 morphological characters that are a combination of the matrices from six major previous studies (Backeljau et al., 1993; Zrzavy et al., 1998; Peterson and Eernisse, 2001; Brusca and Brusca, 2003; Glenner et al., 2004; Schierwater et al., 2009), as we have found that this often influences the SOM node (Neumann et al., 2021a), when it came to comparable genomic datasets from the literature (Ryan et al., 2013; Chang et al., 2015; Whelan et al., 2015; Simion et al., 2017). The morphological data matrix is based on the Com matrix of our earlier publication (Neumann et al., 2021a), but was reviewed to further remove redundancies (Supplementary Data 3). These 51 characters do not include characters within the Placozoa, as the only clear and consistent character separates Polyplacotoma from the other placozoans which was already clear based on genomic differences.
Following our previous exploration of the weighting space across a number of combinations of large morphological and molecular datasets (Neumann et al., 2021a), we here apply a weighting scheme of 50x weights to the 51 morphology characters. While there is no known ideal weight, we previously observed that morphology tended to influence datasets between 10x and 100x weighting. Accordingly, we chose an intermediate 50x weight that resulted in a ratio of morphological to molecular characters of roughly 1:900, which covered a medium ground of the weighting space (Neumann et al., 2021a). Again, this weighting choice is partially subjective, but we believe allows morphology to not be overwhelmed by the massive quantities of sequence data.
A chronogram — i.e. a time-calibrated phylogeny — was estimated using the chronos function in the R package ape (Paradis, 2013; Paradis and Schliep, 2019). Both the relaxed and the correlated models were examined, but only the relaxed model is presented, as the results were similar and we suspect the related model better reflects the lack of calibration points within Placozoa. Maximum and minimum calibration points were set for nodes outside of Placozoa using estimates from TimeTree 5 (Kumar et al., 2022). Again, no fossil-based or alternative calibration points are currently available within Placozoa, making the chronogram output preliminary.
Some genomes were of better quality than others, both in the databases, as well as from our own samples, due to various reasons related to sampling and sequencing methods. To shield the sensitive downstream gene content analyses (i.e., gene presence/absence) among placozoans from inflated absence data from lower-quality genomes, we only used data from genomes with ≥90% BUSCO scores (complete + fragmented). This removed the placozoan lineages H6 and H11, both ctenophores (Mnemiopsis and Pleurobrachia) and the sponge Oscarella.
A parsimony tree was reconstructed using gene content (i.e., presence/absence) via TNT v1.5 (Goloboff and Catalano, 2016) with the following settings: “xmult: replications 1,000 ratchet 5 drift 5 fuse 5.” Bootstraps were run with 1,000 replicates: “resample boot repl 1,000 from 0 [xmult = replications 10 ratchet 5 drift 5 fuse 5]”.
Ancestral state reconstructions
Shifts in gene content across our phylogenomic matrix were principally assessed using ancestral state reconstructions. We used both a continuous data approach and a more nuanced approach looking at each gene’s presence/absence across the tree. The likelihood subtree for Placozoa (minus H6 and H11) was used as the topology. This was done using Polyplacotoma as an outgroup as well as with the full dataset. While ultimately it would be ideal to focus on further outgroups, the orthology methods resulted in a rather Placozoa-centric set of loci, making it inappropriate to focus on shifts in loci from the outgroup compared to the ingroup. Accordingly, we only present the Placozoa-only analyses, as the results were similar for both analyses and we do not wish to have any misleading results.
Continuous trait maximum likelihood ancestral state reconstruction used the fastAnc function in the R package phytools (Revell, 2012). The continuous trait for each terminal was the sum of orthologs it had in the phylogenomic matrix. This analysis is straightforward and easy to interpret, but does not account for the fact that the distribution of each gene across terminals in our matrix is known.
To account for this history of each gene in the matrix, binary ancestral state reconstructions were performed with the ace function in the R package ape (Paradis and Schliep, 2019) using the following settings: “model = ‘ER’” for equal rates and “type = ‘discrete’.” Binary ancestral state reconstructions were performed for each gene in our presence/absence matrix (i.e., reconstructions for 15,849 orthologs). The individual reconstruction outputs were rounded to be present or absent for each gene for each node. The sum of present genes was then depicted as pie charts for each node.
Uniquely present and absent genes
Separately, genes that were uniquely present or absent for a clade or terminal (as compared to the rest of the placozoans within our phylogenomic matrix) were counted. Principal components analysis (PCA) and affinity propagation (Frey and Dueck, 2007) were used to show in additional formats how our genomes grouped and separated from one another. The PCA was conducted with the “prcomp” function in R. Affinity propagation was conducted with the R package apcluster (Bodenhofer et al., 2011) and eponymous function with the following parameters: “negDistMat(r = 2).”
To address the functions of the uniquely present and absent genes for the different placozoan taxa, we carried out functional annotations and checked for functional group enrichment. Functional annotations were performed by converting protein sequence alignments from each ortholog into hidden markov models (HMMs) using hmmer v3.3.2 (Eddy, 2022). Those HMMs were searched against the Uniprot-SwissProt database and the best hit to each HMM with a full sequence e-value <1×10−5 and a single domain e-value <1×10−4. Best hit UIDs were associated with OGs and used to collect GO terms associated to each UID using online resources at uniprot.org (UniProt Consortium, 2021). A gene ontology annotation (.goa) file was created linking each ortholog to its respective GO terms and this was used as the background set for GO term enrichment analysis (15,849 OGs total). Enrichment of GO terms was analyzed using topGO (v2.44.0) (Rahnenfuhrer, 2022) in R.
Results and discussion
Genomes
We more than doubled the total phylogenetically-relevant protein data for sequenced placozoan genomes, rising from six to 13. Most significantly, we provide protein data in our phylogenomic matrix for Polyplacotoma mediterranea, a species that is substantially divergent from all other placozoans with respect to its physical traits and genetics. This helps to root our understanding of this unusual phylum in terms of evolution, and provides a jumping off point for comparative genomic studies across Placozoa.
All of the new placozoan genomes have ~10,000–13,000 orthologous protein sequences (Table 1), which is similar to prior findings (Srivastava et al., 2008; Eitel et al., 2018; Kamm et al., 2018; Laumer et al., 2018). BUSCO scores were reasonably high across our placozoans (92.7% mean score; Table 1). Two BUSCO scores for placozoans from another study (Laumer et al., 2018) were below 90% (79.50 and 84%), but all others >90%. While all data were used for our likelihood tree-building, only genomes with ≥90% BUSCO scores were retained for gene content analyses.
Phylogenomics
Our phylogenomic reconstructions vary in determining the sister group to all other Metazoa (SOM): When analyzing molecular data only, we retrieve Ctenophora as the SOM (Supplementary Figure 1). When analyzing only morphological data, we retrieve Porifera as the SOM (Neumann et al., 2021a,b). In our joint analysis combining the molecular (2.3 million characters) and morphological data (51 morphological characters, upweighted 50 times), Porifera is inferred as the SOM. In this analysis, every morphological character is outweighed by molecular characters 900:1. We suggest morphology is important in systematics, following our prior work (Neumann et al., 2021a), and focus on the combination as our main likelihood tree (Figure 1A). The position of Placozoa, the primary focus of our paper, remains as sister to a Cnidaria+Bilateria clade in both analyses — a result mirroring the findings of prior studies (Dunn and Ryan, 2015; Feuda et al., 2017; Simion et al., 2017).
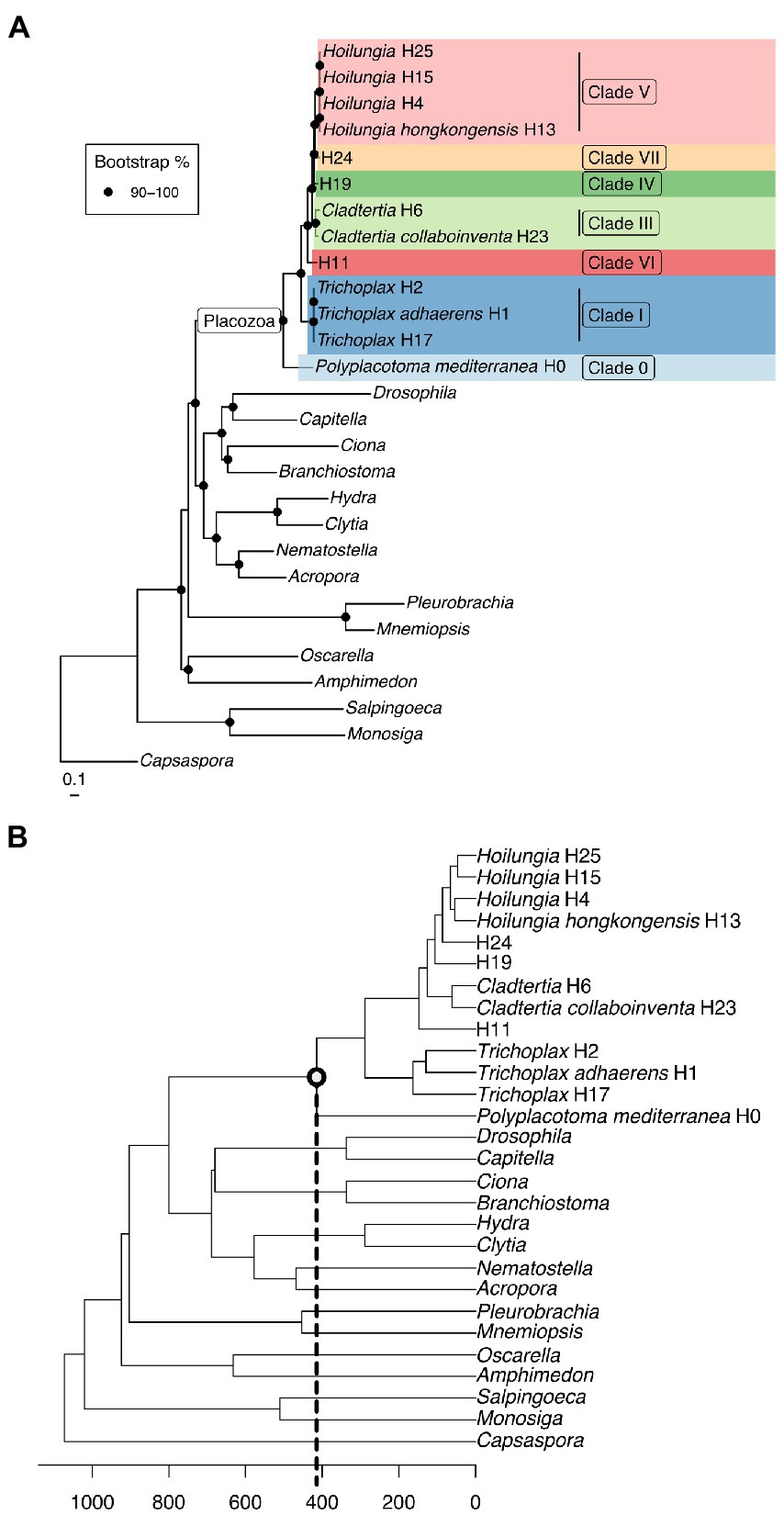
Figure 1. (A) Maximum likelihood phylogenomic tree with most known clades of the phylum Placozoa. It was built using amino acid data from 2,309,771 characters spread across 1,882 orthologs (only those found in ~90% of taxa), plus 51 morphological characters with 50X weighting. Clade nomenclature follows prior research (Eitel et al., 2013), with the addition of Clade 0. This does not include our updated nomenclature for higher taxonomy, but does give our narrower definition of Hoilungia. (B) Chronogram of the maximum likelihood phylogenomic topology (calibration points are based on TimeTree). Please note that the focal node (and all other internal Placozoa nodes) are not fossil calibrated, and accordingly can only be considered a very rough estimate.
Placozoans appear to diversify on a fairly long branch in our likelihood trees (Figure 1), implying that while the phylum is ancient, extant members diversified later — potentially several hundred million years after Placozoa diverged from other Metazoa based on our time calibration (Figure 1B). Still, while placozoan species diversity is more recent in terms of Metazoa evolution, the divergences within Placozoa are old. Our chronogram estimates that the earliest split within Placozoa occured over 400 mya (Figure 1B).
The long branch prior to the diversification of Placozoa can either be explained by: (1) a long period without speciation and a recent onset of genetic radiation or (2) a bottleneck event (the extinction of all but one surviving placozoan lineage), which we regard as far more plausible (Eitel et al., 2018; Laumer et al., 2018). While our time-calibrated tree also suggests that the bauplan of placozoans has remained the same for hundreds of millions of years, they certainly underwent genome evolution between their origin and the more recent radiation (Ax, 1996; Dohrmann and Wörheide, 2013; Ferrier, 2016; Laumer et al., 2018; Schierwater et al., 2021a). Please also note that our time-calibrated tree is surely preliminary in its time scale, as no placozoan fossils at this time can be used to anchor the tree with actual dates. Only very recently has the first putative placozoan fossil been announced (Knaust, 2021). Without having seen these soft-body fossils ourselves, which appear to be of larger body size than any recent placozoans we have observed, we do not intend to make a statement on their suggested placozoan identity. Either way, the date of ~242 myo for the fossil neither confirms nor contradicts our molecular clock analysis.
Relationships within Placozoa are strongly supported in likelihood (almost all 100% bootstrap). Most importantly in terms of relationships amongst placozoans, P. mediterranea is sister to all other phylum members and separated by a reasonably long branch (again, this divergence is estimated at over 400 mya). This is in agreement with previous phylogenetic analyses based on nine mitochondrial protein coding loci as well as two nuclear ribosomal RNA loci (Osigus et al., 2019). Polyplacotoma mediterranea is currently the only species described in its genus, and is the only placozoan species that is easily morphologically distinguished from the other species and lineages: most placozoans are mostly circular and flattened, somewhat like a plate; P. mediterranea, while still ameboid in appearance, is highly and irregularly branched. This confirms the finding that P. mediterranea differs to such a degree that warrants placing it in its own genus (Osigus et al., 2019). Osigus et al. (2019) did not erect a new family for P. mediterranea based on mitochondrial genome as well as nuclear ribosomal RNA data only. However, given the confirmation of its impressive uniqueness we now place it into its own family, order, and class (see below). Please note that the molecules-only tree and the molecules-plus-morphology tree are identical within Placozoa, as the morphology matrix did not include within-Placozoa characters (again, placozoans are morphologically invariant except for P. mediterranea).
Overall, our topology within Placozoa is in strong agreement with a recently published whole mitogenome analysis (Miyazawa et al., 2021). The only points of discordance are with two species-level relationships within the genera Trichoplax and Hoilungia: Within genus Trichoplax (Clade I), we retrieve T. adhaerens as sister to H2, not H17; within genus Hoilungia (Clade V), we retrieve H4 as sister to H13, not H15. We also find the newly sampled lineage H25 in Clade V. Importantly, the similarities of clades and their relationships between our work and the recent mitogenomics results indicate a high degree of stability with datasets using large numbers of loci. The 16S gene tree generally recovers the same named clades, but has more difficulty in distinguishing other relationships, such as those found among clades. This suggests that 16S may continue to be useful for easy identification of haplotypes and clades, but should not be used to build phylogenetic trees across the phylum.
The gene content phylogeny (parsimony using gene presence/absence) was congruent in terms of Placozoa relationships with the likelihood phylogeny that used amino acid data (Figure 2). This, again, suggests that the relationships are relatively stable within this phylum for our dataset.
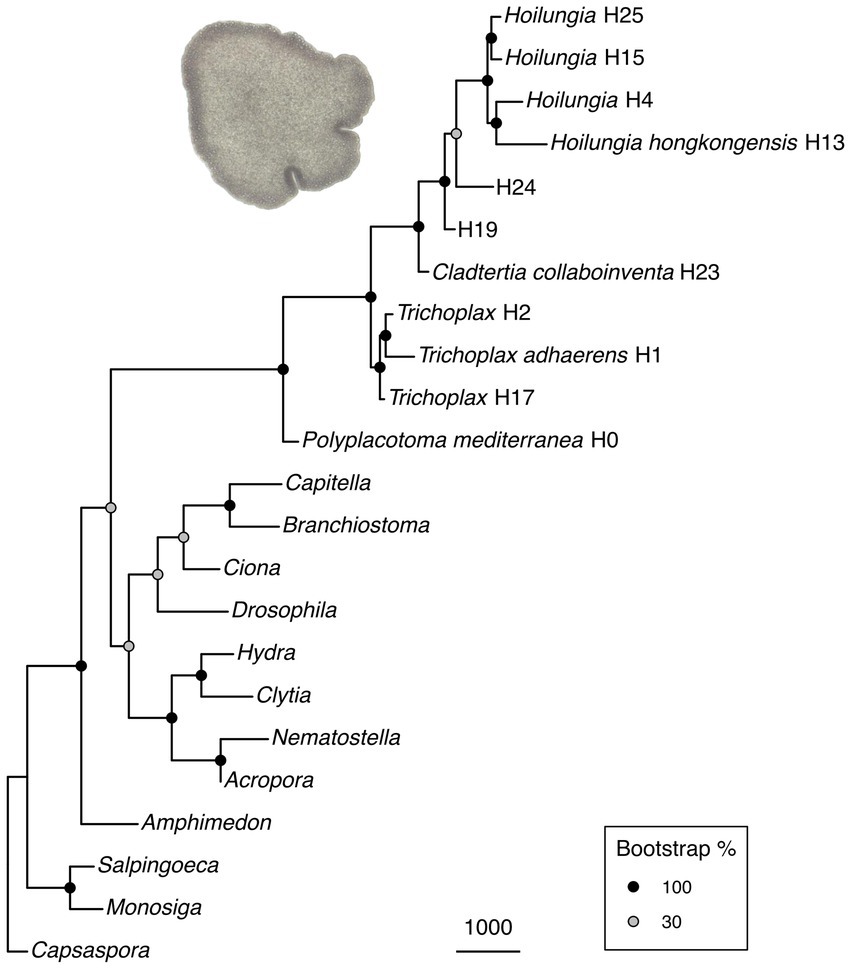
Figure 2. Parsimony reconstruction of most known clades of the phylum Placozoa using gene content (i.e., presence/absence) data for 15,849 genes. Only data from genomes with ≥90% BUSCO scores (complete + fragmented) were used for this analysis to avoid inflated absence data for lower-quality genomes. This removed placozoan lineages H6 and H11, as well as the ctenophores (Mnemiopsis and Pleurobrachia) and a sponge (Oscarella). The image is Trichoplax adhaerens (displays the typical morphology known for Placozoa lineages other than Polyplacotoma).
Gene content
Recent studies investigated gene gains and losses across all animal phyla plus outgroups (Pett et al., 2019; Fernández and Gabaldón, 2020). Across metazoan phyla, there are two trends: The majority of gene gains and duplications happened early inanimal evolution, with phylum-specific gene losses following in more recent evolutionary times (Fernández and Gabaldón, 2020). This suggests that the amount of genes in the repertoire of a lineage does not correlate directly with morphological complexity. Here we present the first genome-wide gene content analysis for the different lineages of the phylum Placozoa.
While placozoans are morphologically similar, we find that their genomes are anything but uniform. Ancestral state reconstructions of gene content (presence/absence) in the placozoan genomes indicate that there have been at least three major shifts towards an increase in the genes in our matrix (Figure 3). The first gain comes about with the clade comprising all placozoans other than P. mediterranea. This once again highlights the uniqueness of P. mediterranea compared to other placozoans, as it does not have 600 orthologs that are found within all other placozoans analyzed (Figure 3). The second gain of genes in our matrix is for the clade excluding P. mediterranea and the genus Trichoplax (the new orders Cladhexea and Hoilungea, see taxonomy section below). The third major gain occurs for the genus Hoilungia (sensu stricto, see taxonomy below; Clade V), which appears to have the largest number of genes present across our matrix.
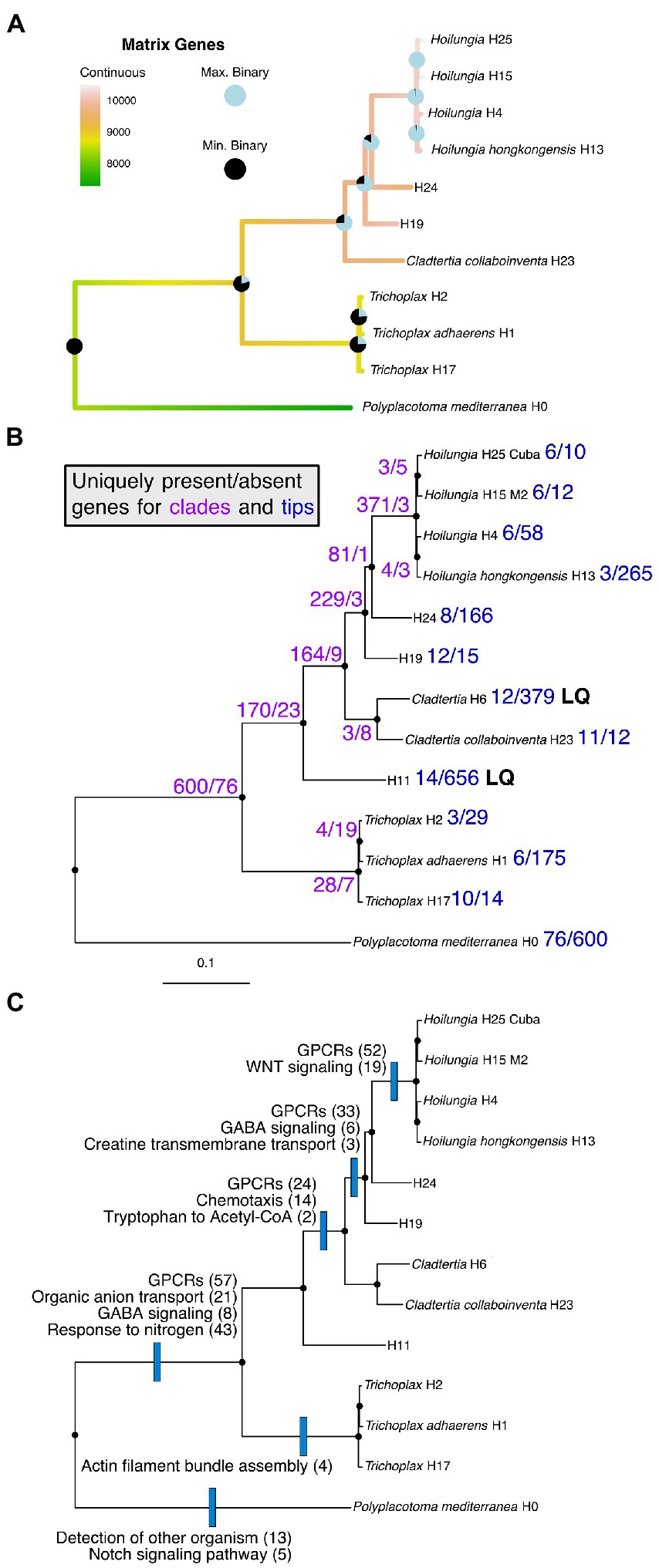
Figure 3. (A) Ancestral state reconstructions (continuous and sum of binary analyses) mapped onto the phylogenomic likelihood subtree containing most known clades of Placozoa. Binary reconstructions were conducted for each of 15,849 genes, and are depicted as pie charts ranging from the minimum number of genes found for any node to the maximum number found for any node. Only data from genomes with ≥90% BUSCO scores (complete + fragmented) were used for this analysis to avoid inflated absence data for lower-quality genomes (other tips were trimmed from the phylogenomic likelihood tree). This removed placozoan lineages H6 and H11. (B) The full Placozoa likelihood subtree with the numbers of uniquely present or uniquely missing genes found for each clade or tip. LQ = low quality (see BUSCO scores in Table 1). (C) The full Placozoa likelihood subtree with the names and numbers of functional genetic groups enriched among uniquely present orthologs. Functional enrichment is based on GO terms.
Please note that gene content (i.e., presence/absence) shifts are only for orthologs in our phylogenomic matrix, rather than the entirety of genes across all genomes we assess. Given the bulk of our genomes are from placozoans, the orthologs appear to be strongly skewed towards Placozoa genes, which we feel allows us to show Placozoa-associated gene shifts over time.
Figure 3B details the clade and terminal specific orthologs, and reveals major shifts at multiple points in Placozoa evolution, such as comparing P. mediterranea to the other major lineage. It also shows major shifts within the former Hoilungia/Group A lineage (based on these findings we reclassify this below). Figure 3C shows a subset of the uniquely present genes found in Figure 3B, and adds information about changes in gene functions based on GO term enrichment. GO term enrichment statistics are also shown in Supplementary Data 4.
The PCA and affinity propagation generally mirrored the major divergence events found with our comparative and phylogenomic analysis (Figure 4): P. mediterranea is clearly divergent, and Trichoplax and Hoilungia are clearly distinctive as well in Figure 4B. Additionally, Figure 4B suggests H19, H24, and Cladtertia collaboinventa H23 are distinct from Trichoplax and Hoilungia. Our new classification below reflects this. Lastly, it is noteworthy that in Figure 4B there is substantial gene content separation within Hoilungia, even though this was less apparent with phylogenomics.
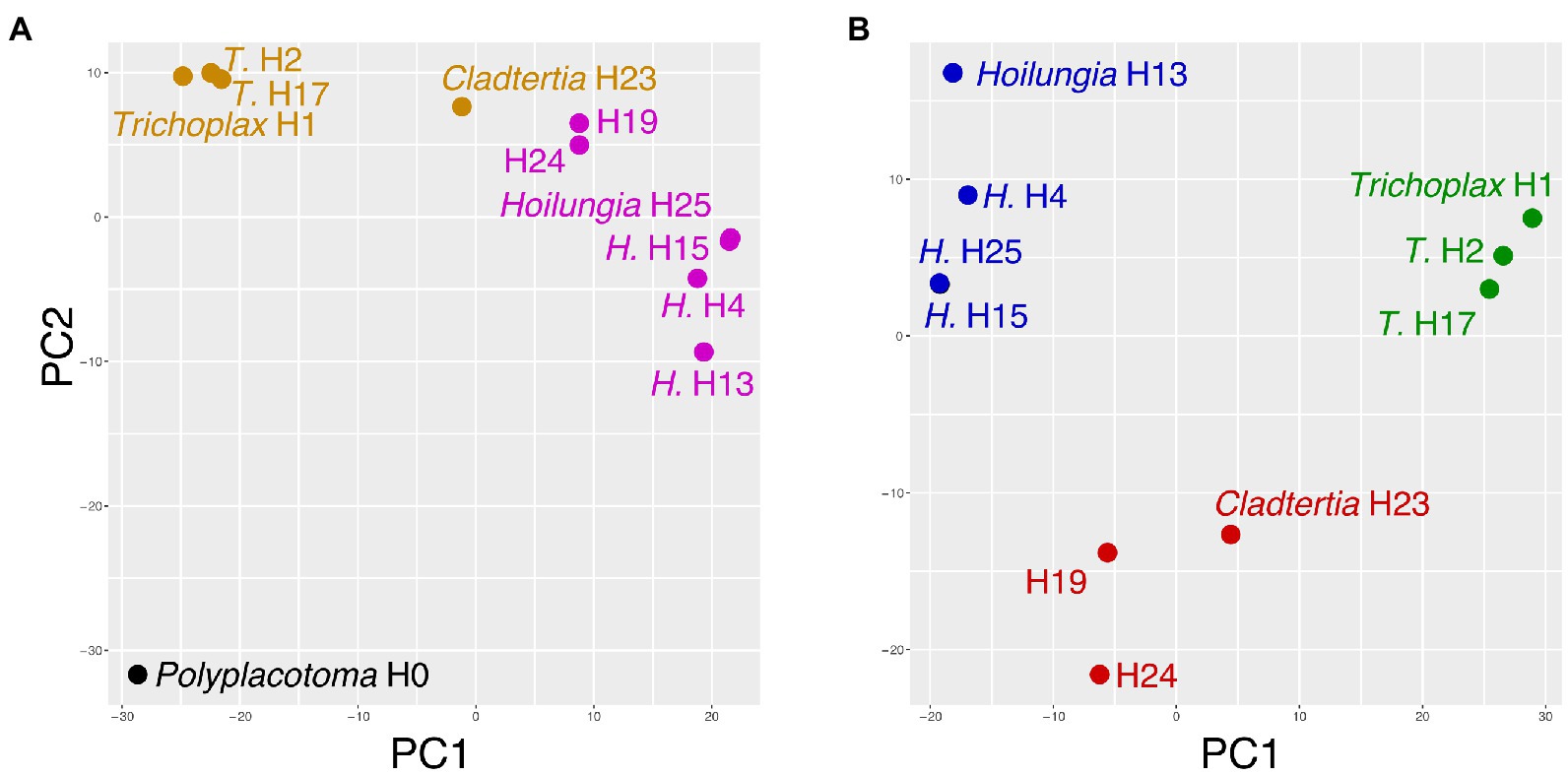
Figure 4. Principal components analysis of gene content (i.e., presence/absence). (A) Includes Polyplacotoma mediterranea (variance explained PC1 = 42%, PC2 = 15%). (B) excludes P. mediterranea (variance explained PC1 = 45%, PC2 = 16%), as it is so divergent from the other placozoans. In both plots, coloring is based on clusters selected using affinity propagation on these two datasets.
Classification background
Our robust phylogenomic reconstructions, along with the last few decades of concordant research on placozoan diversity (Grell, 1971a; Voigt et al., 2004; Eitel et al., 2013, 2018; Paknia and Schierwater, 2015; Osigus et al., 2019), finally puts us in a position to establish a backbone taxonomy. Here we formalize the higher taxonomy within the phylum, providing formal and informative names for the major placozoan lineages, rather than simply referencing them as mathematically numbered clades and haplotypes. Given everything we know about placozoan diversity, the genome-wide genetic differences we report on here reflect broad divergences. The two placozoan genomes (H. hongkongensis and T. adhaerans) that have previously been compared to other metazoans were in the range of genus to order level separation (Eitel et al., 2018). As placozoans in this study are even more divergent, our new classification ranges up to class.
Before detailing our new taxonomy, we consider the background on the state of Placozoa taxonomy. Currently, there are three described genera. Polyplacotoma is monotypic and has so far not been assigned to a higher taxonomic rank such as a family (Osigus et al., 2019). All other placozoan lineages have been grouped into the Trichoplax or Hoilungia, and put into the family Trichoplacidae (Bütschli and Hatschek, 1905). This sums up the prior higher taxonomy within Placozoa, which expanded with the second and third genus + species descriptions in this phylum over the last few years. This is remarkable progress, given that it was over 135 years since the original species and genus Trichoplax was described (Schulze, 1883). However, we believe the prior nomenclature was simply a starting point.
When we placed our data into a phylogenomic and comparative context, it suggested to us that it is time to expand this taxonomy to improve nomenclatural stability and communicability. The single family Trichoplacidae has not been reassessed since the discovery of Polyplacotoma [which, based on our and previous results (Osigus et al., 2019), is highly divergent] or in a phylogenomic context like we present, where most of the major lineages are included. These genera were established based on a comparison of genetic distances in other early-branching animal phyla. That classification was conservative, taking the focus to the genus level, when it was equally possible to expand to the family, or possibly even order level, based on those results. So, we further reviewed the number of higher taxa within other major metazoan lineages (those that have been considered as the potential SOM) to get a sense of whether a broader nomenclature was warranted. The comparison is in Table 2.
Placozoa is the least-studied phylum of the non-bilaterian lineages. It remains impossible to directly observe placozoans in the field, making collecting efforts burdensome. Accordingly, it took over a century to find Polyplacotoma — the only placozoan that looks strikingly different to others with light microscope. In total, this makes us believe we have only discovered the tip of the iceberg in terms of Placozoa species.
Assuming most known 16S haplotypes are equivalent to species, we would have ~25 placozoan species, only three of which have been described. A previous study using a conspecificity matrix found allele sharing for three nuclear-encoded ribosomal proteins between different placozoan haplotypes and suggested several be joined into one species, namely all haplotypes in genus Hoilungia into H. hongkongensis. (This analysis, however, does show that reproductive isolation exists between the placozoan higher level clades, showing strong support for the deep splits in the phylum that we focus on in our higher taxonomy.) Accordingly, the exact number of extant placozoan species is imprecise. Still, we caution against a narrow view of the biological species concept as it often fails to characterize the existing species diversity adequately — even in non-cryptic cases, as exemplified by interbreeding between species like wolves and coyotes (Rutledge et al., 2010; Wheeldon and Patterson, 2012). Of course, further studies must delve deeper into determining the border between placozoan populations and species.
Based on our comparative genomic approach using likelihood phylogenomics and gene content analysis, the 13 placozoans in this study appear to be distinguishable species, and represent members from most major clades within the phylum (Eitel et al., 2013). As the most divergent placozoan was only recently discovered, it is likely that many of the yet-discovered placozoans will represent new higher-level taxonomic groups.
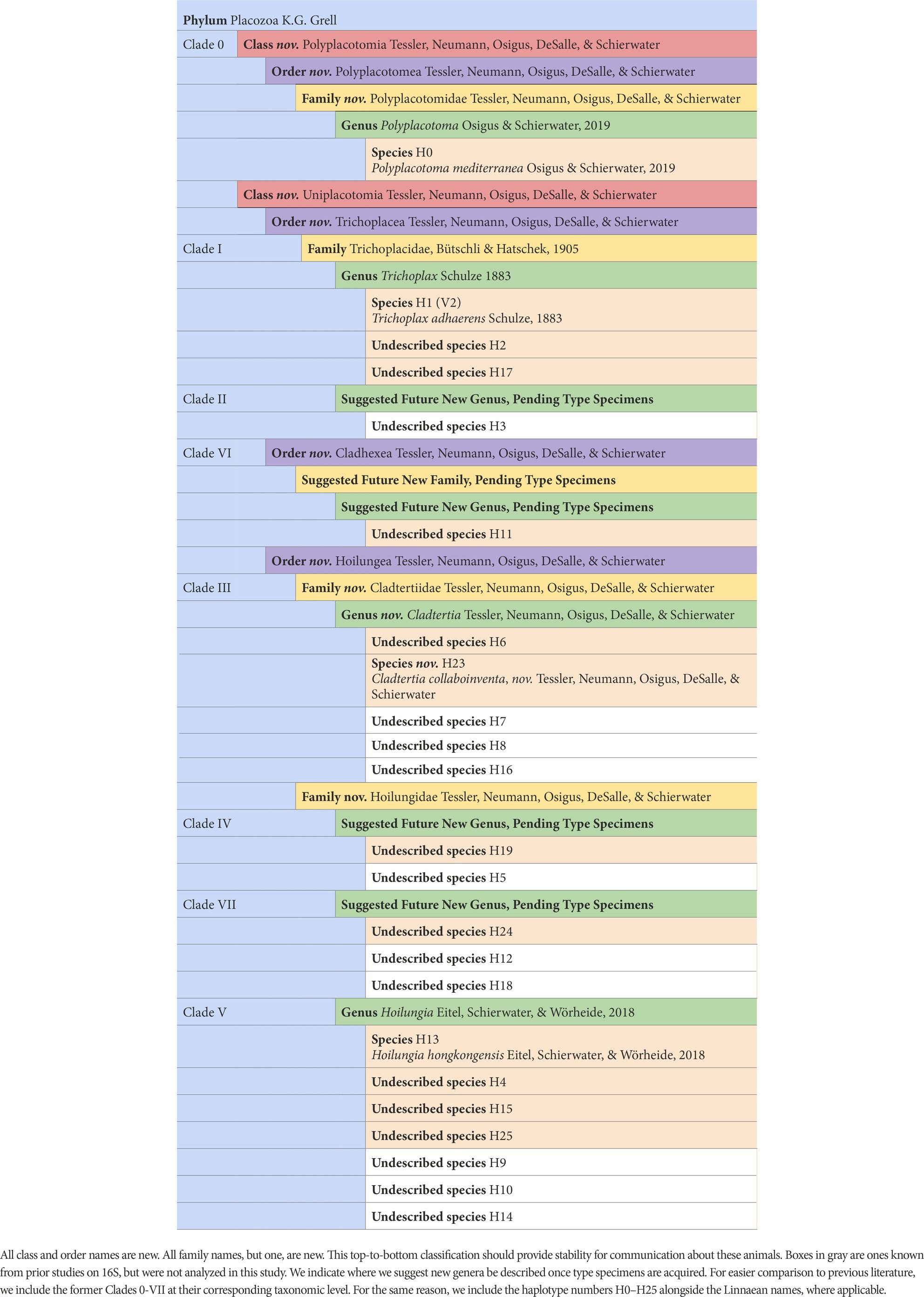
It has already been estimated, albeit somewhat speculatively, that there are likely 100’s of placozoan species (Eitel and Schierwater, 2010). Ctenophores have 2 classes, 9 orders, 30 families, 55 genera, and ~ 200 described species. The ratio of higher-level taxonomic groups to described species is greater for Ctenophora than for Cnidaria or Porifera, but as the closest analog to Placozoa in terms of species diversity, having a similar ratio seems within reason (Table 2). Accordingly, we have increased the number of higher taxa within Placozoa substantially (Table 3 and Figure 5).
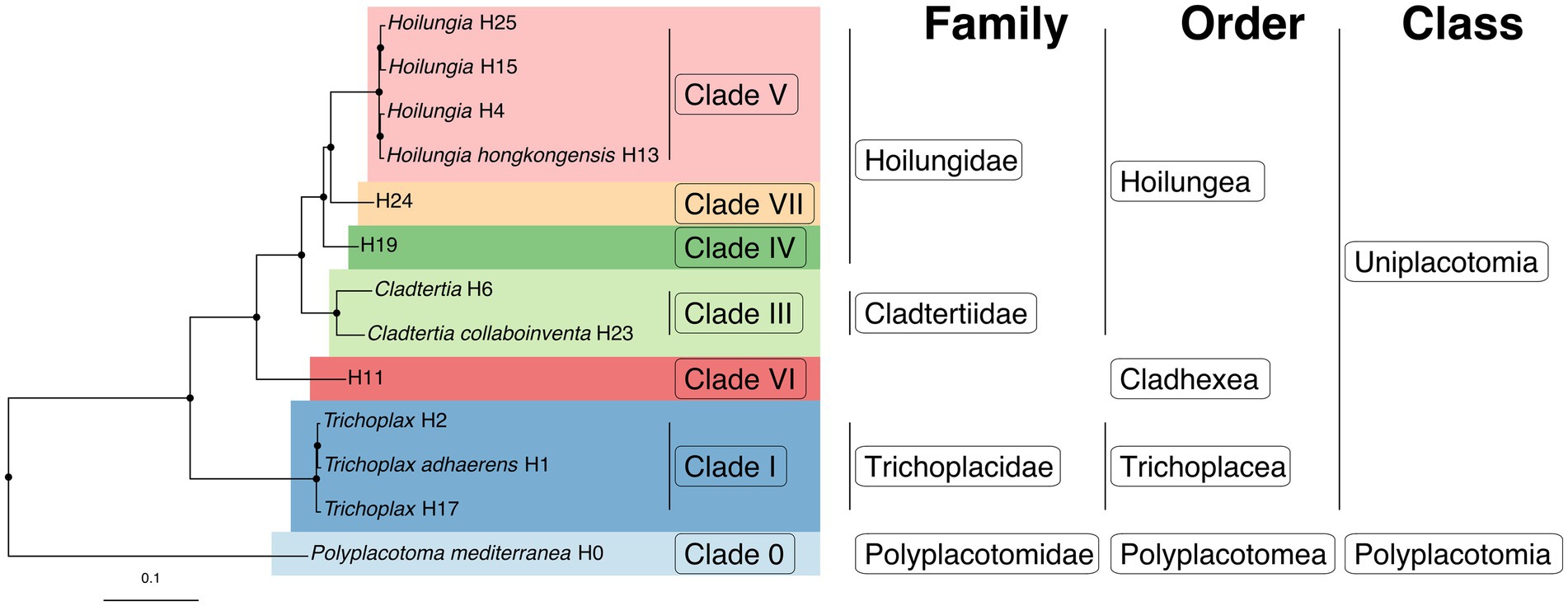
Figure 5. The first fully fleshed-out taxonomy for the phylum Placozoa displayed on the likelihood phylogenomic subtree (also see Table 3). Please note that around twice as many species-level lineages have been discovered that are yet to be incorporated into a phylogenomic tree.
Complex genome structural changes appear to be an effective method for describing placozoan species (Eitel et al., 2018). This seems like a highly promising avenue forward, but it requires much more sequencing and computational resources [Hi-C is likely to be of aid (Belton et al., 2012)]. In contrast, gene content analyses provide another method for finally dealing with placozoan taxonomy that is straightforward and assessable with a variety of sequencing techniques.
Gene content makes for easy Placozoa taxonomy. The existence of one or more clade-specific genes can identify species or higher taxa. We suggest using multiple clade-specific genes for higher precision, because genome sequencing may miss a gene on occasion or copy number might vary. Of course, some traditional biologists prefer basic morphology (physical features that are in some way visualizable) for taxonomy, but this will likely never be a proper solution for Placozoa or other groups with simple morphology (e.g., most bacteria). Still, gene content is character-focused, and accordingly more congruent with the practices used by most systematists while describing a species using morphology. In fact, we believe gene content is simply a form of molecular morphology. The use of this type of molecular morphology is discussed in further detail in Tessler et al. (2022). Furthermore, given that we and others have argued that even single nucleotides can be sufficient for delineating species (DeSalle et al., 2005), then the presence/absence of an entire gene (or several as we suggest) should be a similar but stronger way towards classification. It is worth noting that what is believed to be clade-specific currently will change to some degree with further sequencing of genomes, as some genes may be more widespread than initially thought. However, that is not different in principle to other methodologies, as the utility of taxonomic characters is always refined with further sampling.
There is one noteworthy problem with the technique. If using full genome assemblies, there can be issues with poor-quality genomes. This problem is exclusively for absence of genes rather than presence of genes. Accordingly, for poor-quality genomes, focusing on gene presence is the best diagnostic.
Lastly, while the purpose of our gene content analysis is simply to compare orthologs as presence/absence characters (Figure 3C), highlights functional enrichment among uniquely present orthologs. Functional enrichment of sets of uniquely present orthologs at different nodes in the tree is suggestive of the processes underlying the divergence of the different lineages. For example, changes in G-protein coupled receptors (GPRCs) are notable at the base of class Uniplacotomia, order Hoilungea, and family Hoilungidae. Expansion of GPRCs has been noted as a feature of metazoan evolution for their roles in cell–cell communication, developmental control, and environmental sensing (de Mendoza et al., 2014). In the case of genus Hoilungia, functional enrichment is refined to WNT signaling, a specific subset of G-protein signaling used during development in other animals (Komiya and Habas, 2008) suggestive of developmental differences between Hoilungia and other placozoans. Overall, the enrichment of functional processes at nodes in the tree adds weight behind these ortholog presence/absence characters, the functions of those orthologs are not randomly distributed, and this surely is fodder for interesting work to be done on the evolutionary shifts that have occured within Placozoa.
First fleshed-out Placozoa taxonomy
Given the benefits detailed above, we primarily use a combination of monophyly, phylogenomic branch length, and gene content analyses (ancestral state reconstructions, uniquely present/absent genes, PCA, and affinity propagation) to establish the separations between our new higher taxa.
We start with only three described genera and species plus a family, as well as informally named clades, but otherwise, as far as Linnaean taxonomy goes, we have a blank slate or tabula rasa for organizing all known placozoan diversity. This is an exceptional moment to classify an entire animal phylum. We list the steps we took here, while the actual classifications are detailed below. The full new taxonomy is found in Table 3 and Figure 5.
Our tree was highly supported and, combined with trans-study-congruence, the implied stability in the monophyletic clades we recovered represents our first requirement for naming.
The next step was to make major divisions within the phylum, and it was clear in every analysis that Polyplacotoma was highly divergent from the rest of the lineages: it is missing 600 orthologs found across the other lineages, comes out as a unique cluster in affinity propagation, and is clearly divergent with phylogenetic topology and PCA distances. Considering these results and this species’ distinct morphology, we considered this to be a new class: Polyplacotomia class nov. We place all other Placozoa into the new class Uniplacotomia class nov.
Next, we established new orders. Each genomic analysis showed that Trichoplax is highly divergent from the remaining lineages. Accordingly, it seemed prudent to us to give it a unique order, Trichoplacea ord. nov. Our ancestral state reconstruction suggests that the remaining clade underwent a major shift in loci (Figure 3). H11 was not included in gene content analyses, because the genome was less complete, but it is vastly different from the class based on phylogenomics. Therefore, we put in the new order Cladhexea ord. nov. All remaining placozoans form a clade with 164 unique loci, and are thus joined into the new order Hoilungea ord. nov. Again, Polyplacotoma is put into its own new order, Polyplacotomea ord. nov.
Affinity propagation and PCA suggest that we should break up the classification further, particularly within the order Hoilungea ord. nov. Affinity propagation suggests H. hongkongensis, H4, H15, and H25 form a unique cluster. This is mirrored by this clade having 371 unique loci. Accordingly, we believe these should be considered as the only members of the genus Hoilungia, and we therefore revise the members of the genus Hoilungia sensu Eitel, Schierwater & Wörheide (Eitel et al., 2018).
The more difficult question is whether or not this should represent a unique family. Prior 16S analysis suggests that Clade VII (including H24) and Clade IV (including H19) have multiple species (see Table 3) (Eitel et al., 2013). Future phylogenomic studies will need to clarify the exact relationships among these other already discovered species. Our phylogenomic analyses are limited to one terminal each for these, and we aim to avoid the creation of overly split groups (e.g., many families each with one genus). Therefore, we have come to the conclusion that H24 and H19 should eventually represent their own genera, joined with Hoilungia in the family Hoilungidae fam. nov.; unfortunately, we cannot describe them at this point because we do not have material for designating type specimens. Together, the members of this new family share a high number of uniquely present orthologs (229). More data on these understudied placozoan lineages might warrant raising these genera to the family level at a later time, especially given the marked shift in gene content to the genus Hoilungia retrieved in all analyses. Given our creation of the family Hoilungidae and a desire to create as few additional families as possible, while retaining monophyly for them, we erect Cladtertiidae fam. nov. with a new genus Cladtertia gen. nov. This new genus contains a species we describe new here as Cladtertia collaboinventa nov. (H23). This new species is morphologically identical to all other known placozoan lineages except for P. mediterranea when observed via light microscopy (Supplementary Figure 2), but clearly unique when considering its genome. It has 11 genes that are not found in any other placozoans, and lacks 12 genes that all other placozoan genomes contain (Supplementary Datas 5, 6). It was found in a sea water aquarium in Germany and is thus of unknown geographical origin (see Table 1). The genus further contains H6, which remains undescribed due to lack of material to archive as type specimens. Trichoplax remains in Trichoplacidae and Polyplacotoma is put into the new family Polyplacotomidae fam. nov. Additionally, the one new species is the minimum amount needed to describe our new higher taxa.
As mentioned, the naming of additional species, genera, and one new family as listed in Table 3 was not possible in this study. This is because the International Commission on Zoological Nomenclature (ICZN) requires a type species with a type specimen for this to be achieved. We do not have the material to designate type specimens for these lineages at this point, but plan to describe these species in future work. Still, we include in Table 3 the areas where we suggest additional names be added. This includes new genera for Clade IV and Clade VII, as well as a new genus and family for Clade VI. Similarly, we look forward to describing the species that we are aware of based on 16S haplotypes with our gene content methods; however, we will similarly need fresh collections to provide type material for these animals. Given our knowledge of these undescribed species, it will be easy to integrate them into the updated taxonomy we present here once we get type material and genome sequences to fully describe them.
Unique gene content was used to diagnose each new taxon (see Supplementary Datas 5, 6). To be explicit, these are genes that are uniquely and consistently present or absent for a taxon rather than something generated by ancestral state reconstructions. This makes them more akin to how morphology is traditionally examined, such as a taxon has a character or it does not. These are unique genes compared to the other Placozoa analyzed thus far. This is not a comparison to non-placozoans. A breakdown detailing our new taxa follows. Please note that we rely on multiple uniquely present and uniquely absent genes for each taxon described to help minimize the chances that one of these genes is the result of an assembly or sequencing issue.
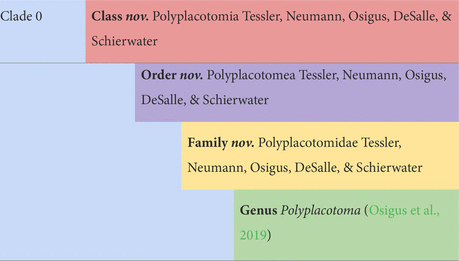
Diagnosis: 76 uniquely present and 600 uniquely missing genes (Supplementary Datas 5, 6). The only highly ramified (multiple elongate, ameboid projections) Placozoa.
Notes: Currently monotypic and the earliest-branching placozoan taxon.
Etymology: Ancient Greek: polús = many; plakós = plate; −toma = that cuts.
Type species: Polyplacotoma mediterranea (Osigus et al., 2019).
Zoobank LSID, Family only: urn:lsid:zoobank.org:act:C3EBFFB7-C044-4588-8A0B-2C56A4182A99.

Diagnosis: 600 uniquely present and 76 uniquely missing genes (Supplementary Datas 5, 6). Ameboid morphology; rarely branching.
Notes: This is the vast majority of known Placozoa lineages (everything other than Polyplacotoma mediterranea).
Etymology: Latin: unus = one; Ancient Greek: plakós = plate; −toma = that cuts.
Type order: Trichoplacea nov. Tessler, Neumann, Osigus, DeSalle, & Schierwater.

Diagnosis: This follows Trichoplax.
Notes: Includes Trichoplax.
Etymology: Latin: Trich = hair, Ancient Greek: plakós = plate.
Type family: Trichoplacidae (Bütschli and Hatschek, 1905).
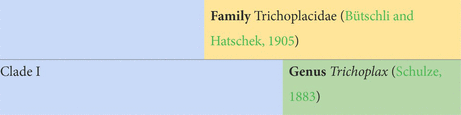
Diagnosis: 28 uniquely present and 7 uniquely missing genes (Supplementary Datas 5, 6). Members share alleles for nuclear-encoded ribosomal proteins rpl9, rpl32, and rpp1 (Eitel et al., 2018). Members have the northern- and southernmost confirmed distributions and predicted habitats of all discovered placozoans.
Notes: Includes T. adhaerens (H1), H2, and H17.
Etymology: Latin: Trich = hair, Ancient Greek: plakós = plate.
Type species: Trichoplax adhaerens (Schulze, 1883).
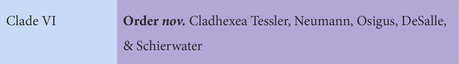
Diagnosis: 14 uniquely present and 656 uniquely missing genes (Supplementary Datas 5, 6).
Notes: Includes H11.
Etymology: Ancient Greek: kládos = branch; hex = 6.

Diagnosis: 164 uniquely present and 9 uniquely missing genes (Supplementary Datas 5, 6).
Notes: Contains most placozoan haplotypes discovered so far.
Etymology: Cantonese: Hoi Lung = Sea dragon.
Type family: Hoilungidae nov. Tessler, Neumann, Osigus, DeSalle, & Schierwater.

Diagnosis: 3 uniquely present (this would surely be higher if we had a better quality genome for H6) and 8 uniquely missing genes (Supplementary Datas 5, 6). Members share alleles for nuclear-encoded ribosomal proteins rpl9, rpl32, and rpp1 (Eitel et al., 2018).
Notes: Includes H6 and Cladtertia collaboinventa H23, and, based on 16S, H8 and H16.
Etymology: Ancient Greek: kládos = branch; Latin: tertius = the third.
Type species: Cladtertia collaboinventa nov. Tessler, Neumann, Osigus, DeSalle, & Schierwater.
ZooBank LSID, Family: urn:lsid:zoobank.org:act:CCD39AD5-CC2E-4E4B-A4E7-BDBFF9831C9B.
ZooBank LSID, Genus: urn:lsid:zoobank.org:act:05A49236-74 BC-4597-81A7-25AA9D6ECD67.

Diagnosis: 11 uniquely present and 12 uniquely missing genes (Supplementary Datas 5, 6).
Notes: This was found in a sea water aquarium.
Etymology: Ancient Greek: kládos = branch, tertia = the third, collaboratio = collaboration, inventa = found, meaning: The third clade that has been found in collaboration, as it required strong altruistic collaborations between lots of partners instead of a single scientist.
Holotype: Specimen AMNH_IZC 00382827 deposited at the American Museum of Natural History.
Paratypes: Specimens AMNH_IZC 00382828, AMNH_IZC 00382829 deposited at the American Museum of Natural History.
ZooBank LSID: urn:lsid:zoobank.org:act:AEA0C36A-1D74-43FC-9F15-7490E3E48313.

Diagnosis: 229 uniquely present and 3 uniquely missing genes (Supplementary Datas 5, 6); also see Eitel et al. (2018).
Notes: Includes Hoilungia hongkongensis (H13), H4, H15, H25, H19 and H24; and further H5, H9, H10, H12, H14, and H18 based on 16S.
Etymology: Cantonese: Hoi Lung = sea dragon.
Type genus: Hoilungia Eitel, Schierwater & Wörheide, 2018 (Eitel et al., 2018).
ZooBank LSID: urn:lsid:zoobank.org:act:782A45D2-41CA-464A-93BB-85EA6AF47246.

Diagnosis: 371 uniquely present and 3 uniquely missing genes (Supplementary Datas 5, 6). All members of this genus share alleles for nuclear-encoded ribosomal proteins rpl9, rpl32, and rpp1 (Eitel et al., 2018).
Notes: Includes Hoilungia hongkongensis (H13), H4, H15, and H25; and further H9, H10, and H14 based on 16S. This is a narrower usage of the Hoilungia than was originally described.
Etymology: Cantonese: Hoi Lung = sea dragon.
Type species: Hoilungia hongkongensis Eitel, Schierwater & Wörheide, 2018 (Eitel et al., 2018).
Our reliance on phylogenetics is in line with PhyloCode (de Queiroz and Cantino, 2020), and we respect that system. Still, we prefer our phylogenetically informed Linnaean-style taxonomy, which fits with nearly all animal classifications and accordingly should promote nomenclatural longevity.
Making this new taxonomy, we believe Placozoa will finally be at a generally more comparable level with the other animals (Table 2), although there is much variability in numbers of higher taxa across animal phyla. This will also help with communication, as Linnaean ranks are taught to school children and clearly indicate levels of nestedness among taxa. Our taxonomy will also allow efficient incorporation of newly discovered lineages, which we fully anticipate given that much more cryptic diversity is surely to be described for one of the oldest animal phyla, as it has a world-wide distribution spanning habitats from clear reefs to brackish waters.
Conclusion
Our study represents the first and potentially only time that a complete Linnaean taxonomy is formally erected for an entire animal phylum. This was powered by our work recovering stable relationships among major placozoan lineages using phylogenomics that matched recent mitogenomics (Miyazawa et al., 2021). The higher-level Linnaean taxonomy is based on gene content analyses combined with our tree-based approaches, which we believe will further improve communication about these animals. Our gene content methods for producing this new taxonomy may also be applicable towards producing stability for other morphologically cryptic but genetically highly variable lineages such as microbes. Our study may also serve as a template to revisit systematics of some other critical metazoan phyla with a new integrative tool set of the 21st century. Lastly, we once again point out the importance of morphology in phylogenomics and the instability of the SOM (sister to other Metazoa, i.e., animals) node (Neumann et al., 2021a), and we lend support towards sponges as the SOM.
Data availability statement
The data presented in the study is deposited as Supplementary material to this paper and at the link: https://doi.org/10.5061/dryad.7m0cfxpz4. Further queries can be directed to the corresponding author.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
JN thanks the DAAD PhD student fellowship and tuition scholarship for supporting part of this project. JB was supported in part by NSF OIA-1826734. H-JO acknowledges a doctoral fellowship of the Studienstiftung des deutschen Volkes. We thank the US DOE (BER Grant DE-SC0014377) awarded to RD for helping to fund this research. This Open Access publication was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - 491094227 “Open Access Publication Funding” and the University of Veterinary Medicine Hannover, Foundation.
Acknowledgments
We would like to thank Ulrike and Günter Oberjatzas for kindly providing live specimens of Cladtertia collaboinventa (H23). Specimens of haplotype H17 have kindly been provided by Dr. Hiroyuki Kaneko and Dr. Ritsu Kuraishi (Research and Education Center for Natural Sciences, Keio University, 4-1-1 Hiyoshi, Kohoku-ku, Yokohama, Kanagawa 223-8521 Japan). We thank Dr. Michael Eitel for sampling haplotype H15 “M2/11” in Hong Kong (22.352728N 114.251733E) in 2012, and for establishing its original culture. We thank Danielle de Carle for advice on an earlier draft of the manuscript, as well as Jonas Keunecke for photographing specimens of Placozoa used with the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.1016357/full#supplementary-material
SUPPLEMENTARY TABLE 1
Information on the metazoan and non-metazoan outgroups.
SUPPLEMENTARY TABLE 2
Genome statistics and BUSCO scores for the placozoan genomes, with BUSCO scores above our cutoff of 90% highlighted in green.
SUPPLEMENTARY DATA 1
Phylogenomic amino acid matrix, as both “.nex” and “.phy” files.
SUPPLEMENTARY DATA 2
Table with all orthologs, a gene content (presence/absence) filter column, consensus sequence, and functional annotations. The file contains a list of all OGs (Column B, “OG_name”) along with a consensus sequence representative of the protein alignment that defines each OG (Column D, “Consensus Sequence”). The file also contains annotation information for each OG including a UniProt identifier (UID) of the best hit UID for each OG (Column E, “UID-annotation”), as well as information associated with that UID such as entry name, protein name, gene name, and all gene ontology IDs linked to the UID (Columns F-I). In addition, Column C (“Placozoa_Unique_PresAbs”) contains information on OGs that are uniquely present or uniquely missing for different clades and tips in the placozoan tree. The data in column C can be used as a filter to select those OGs and their annotations for further analyses of OGs and their functions that may have been important during placozoan evolution.
SUPPLEMENTARY DATA 3
Morphology matrix.
SUPPLEMENTARY DATA 4
Tables containing GO term enrichment statistics for all three GO categories (BP: Biological Process, MF: Molecular Function, and CC: Cellular Component) for the uniquely present/absent orthologs at all major taxonomic splits, as well as for the new species Cladtertia collaboinventa.
SUPPLEMENTARY DATA 5
Unique present and unique missing genes for each taxon classified in our taxonomy. The gene identifiers correspond to column 2 of Supplementary Data 2.
SUPPLEMENTARY DATA 6
Gene content matrix (i.e. presence/absence) matrix as a CSV file. Only data from genomes with ≥ 90% BUSCO scores (complete + fragmented) were used for this, as they were what was used to make the parsimony tree and do the ancestral state analyses.
SUPPLEMENTARY FIGURE S1
Maximum likelihood phylogenomic tree with most known clades of the phylum Placozoa. It was built using only amino acid data from 2,309,771 characters spread across 1,882 orthologs (only those found in ~90% of taxa), without morphological characters.
SUPPLEMENTARY FIGURE S2
Light microscopy photographs of the seven placozoan lineages currently in culture. These photos were taken under identical conditions, settings, and scale by Jonas Keunecke. In addition, we reproduce a previously published photo of H0 Polyplacotoma mediterranea, which is extinct in culture, with permission by Elsevier under license number 5400220990285.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ax, P. (1996). Multicellular Animals. A New Approach to the Phylogenetic Order in nature. Heidelberg: Springer-Verlag.
Backeljau, T., Winnepenninckx, B., and De Bruyn, L. (1993). Cladistic analysis of metazoan relationships: a reappraisal. Cladistics 9, 167–181. doi: 10.1111/j.1096-0031.1993.tb00216.x
Belton, J.-M., McCord, R. P., Gibcus, J. H., Naumova, N., Zhan, Y., and Dekker, J. (2012). Hi–C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276. doi: 10.1016/j.ymeth.2012.05.001
Bodenhofer, U., Kothmeier, A., and Hochreiter, S. (2011). APCluster: an R package for affinity propagation clustering. Bioinformatics 27, 2463–2464. doi: 10.1093/bioinformatics/btr406
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bütschli, O., and Hatschek, S. (1905). Zoologisches Zentralblatt. ed. A. Schuberg. Leipzig: Verlag von Wilhelm Engelmann.
Campbell, M. S., Holt, C., Moore, B., and Yandell, M. (2014). Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinformatics 48:4.11.1–39. doi: 10.1002/0471250953.bi0411s48
Carr, M., Leadbeater, B. S. C., Hassan, R., Nelson, M., and Baldauf, S. L. (2008). Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc. Natl. Acad. Sci. U. S. A. 105, 16641–16646. doi: 10.1073/pnas.0801667105
Chang, E. S., Neuhof, M., Rubinstein, N. D., Diamant, A., Philippe, H., Huchon, D., et al. (2015). Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl. Acad. Sci. U. S. A. 112, 14912–14917. doi: 10.1073/pnas.1511468112
Chiu, J. C., Lee, E. K., Egan, M. G., Sarkar, I. N., Coruzzi, G. M., and DeSalle, R. (2006). OrthologID: automation of genome-scale ortholog identification within a parsimony framework. Bioinformatics 22, 699–707. doi: 10.1093/bioinformatics/btk040
Daly, M., Brugler, M. R., Cartwright, P., Collins, A. G., Dawson, M. N., Fautin, D. G., et al. (2007). The phylum Cnidaria: a review of phylogenetic patterns and diversity 300 years after Linnaeus*. Zootaxa 1668, 127–182. doi: 10.11646/zootaxa.1668.1.11
de Mendoza, A., Sebé-Pedrós, A., and Ruiz-Trillo, I. (2014). The evolution of the GPCR signaling system in eukaryotes: modularity, conservation, and the transition to metazoan multicellularity. Genome Biol. Evol. 6, 606–619. doi: 10.1093/gbe/evu038
de Queiroz, K., and Cantino, P. D. (2020). International Code of Phylogenetic Nomenclature (PhyloCode): A Phylogenetic Code of Biological Nomenclature. Boca Raton CRC Press.
Dellaporta, S. L., Xu, A., Sagasser, S., Jakob, W., Moreno, M. A., Buss, L. W., et al. (2006). Mitochondrial genome of Trichoplax adhaerens supports placozoa as the basal lower metazoan phylum. Proc. Natl. Acad. Sci. U. S. A. 103, 8751–8756. doi: 10.1073/pnas.0602076103
DeSalle, R., Egan, M. G., and Siddall, M. (2005). The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1905–1916. doi: 10.1098/rstb.2005.1722
DeSalle, R., Tessler, M., and Rosenfeld, J. (2020). Phylogenomics: A Primer. 2nd Edn. Boca Raton: CRC Press.
Dohrmann, M., and Wörheide, G. (2013). Novel scenarios of early animal evolution--is it time to rewrite textbooks? Integr. Comp. Biol. 53, 503–511. doi: 10.1093/icb/ict008
Dunn, C. W., and Ryan, J. F. (2015). The evolution of animal genomes. Curr. Opin. Genet. Dev. 35, 25–32. doi: 10.1016/j.gde.2015.08.006
Eddy, S. R. (2022). HMMER, version 3.3.2 Available at:.hmmer.org (Accessed 01 October, 2022).
Eitel, M., Francis, W. R., Varoqueaux, F., Daraspe, J., Osigus, H.-J., Krebs, S., et al. (2018). Comparative genomics and the nature of placozoan species. PLoS Biol. 16:e2005359. doi: 10.1371/journal.pbio.2005359
Eitel, M., Guidi, L., Hadrys, H., Balsamo, M., and Schierwater, B. (2011). New insights into placozoan sexual reproduction and development. PLoS One 6:e19639. doi: 10.1371/journal.pone.0019639
Eitel, M., Osigus, H.-J., DeSalle, R., and Schierwater, B. (2013). Global diversity of the Placozoa. PLoS One 8:e57131. doi: 10.1371/journal.pone.0057131
Eitel, M., and Schierwater, B. (2010). The phylogeography of the Placozoa suggests a taxon-rich phylum in tropical and subtropical waters. Mol. Ecol. 19, 2315–2327. doi: 10.1111/j.1365-294X.2010.04617.x
Enright, A. J., Van Dongen, S., and Ouzounis, C. A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584. doi: 10.1093/nar/30.7.1575
Erpenbeck, D., Voigt, O., Adamski, M., Adamska, M., Hooper, J. N. A., Wörheide, G., et al. (2007). Mitochondrial diversity of early-branching metazoa is revealed by the complete mt genome of a haplosclerid demosponge. Mol. Biol. Evol. 24, 19–22. doi: 10.1093/molbev/msl154
Feng, J.-M., Xiong, J., Zhang, J.-Y., Yang, Y.-L., Yao, B., Zhou, Z.-G., et al. (2014). New phylogenomic and comparative analyses provide corroborating evidence that Myxozoa is Cnidaria. Mol. Phylogenet. Evol. 81, 10–18. doi: 10.1016/j.ympev.2014.08.016
Fernández, R., and Gabaldón, T. (2020). Gene gain and loss across the metazoan tree of life. Nat Ecol Evol. 4, 524–533. doi: 10.1038/s41559-019-1069-x
Ferrier, D. E. K. (2016). The origin of the Hox/ParaHox genes, the ghost locus hypothesis and the complexity of the first animal. Brief. Funct. Genomics 15, 333–341. doi: 10.1093/bfgp/elv056
Feuda, R., Dohrmann, M., Pett, W., Philippe, H., Rota-Stabelli, O., Lartillot, N., et al. (2017). Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr. Biol. 27, 3864–3870.e4. doi: 10.1016/j.cub.2017.11.008
Frey, B. J., and Dueck, D. (2007). Clustering by passing messages between data points. Science 315, 972–976. doi: 10.1126/science.1136800
Funch, P., and Kristensen, R. M. (1995). Cycliophora is a new phylum with affinities to Entoprocta and Ectoprocta. Nature 378, 711–714. doi: 10.1038/378711a0
Gazave, E., Lapébie, P., Renard, E., Vacelet, J., Rocher, C., Ereskovsky, A. V., et al. (2010). Molecular phylogeny restores the supra-generic subdivision of homoscleromorph sponges (Porifera, Homoscleromorpha). PLoS One 5:e14290. doi: 10.1371/journal.pone.0014290
Glenner, H., Hansen, A. J., Sørensen, M. V., Ronquist, F., Huelsenbeck, J. P., and Willerslev, E. (2004). Bayesian inference of the metazoan phylogeny; a combined molecular and morphological approach. Curr. Biol. 14, 1644–1649. doi: 10.1016/j.cub.2004.09.027
Goloboff, P. A., and Catalano, S. A. (2016). TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 32, 221–238. doi: 10.1111/cla.12160
Goloboff, P. A., Farris, J. S., and Nixon, K. C. (2008). TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786. doi: 10.1111/j.1096-0031.2008.00217.x
Grell, K. G. (1971a). Trichoplax adhaerens F.E. Schulze und die Entstehung der Metazoen. Naturwissenschaftliche Rundschau 24, 160–161.
Grell, K. G. (1971b). Embryonalentwicklung bei Trichoplax adhaerens F. E. Schulze. Die Naturwissenschaften 58:570. doi: 10.1007/BF00598728
Grell, K. G. (1972). Eibildung und Furchung von Trichoplax adhaerens F. E. Schulze (Placozoa). Zeitschrift für Morphologie der Tiere 73, 297–314. doi: 10.1007/BF00391925
Grell, K. G., and Benwitz, G. (1971). Die Ultrastruktur von Trichoplax adhaerens F.E. Schulze. Cytobiologie 4, 216–240.
Grell, K. G., and Benwitz, G. (1974). Spezifische Verbindungsstrukturen der Faserzellen von Trichoplax adhaerens F. E. Schulze. Zeitschrift für Naturforschung C 29, 790–790b.
Grell, K. G., and Benwitz, G. (1981). Ergänzende Untersuchungen zur Ultrastruktur von Trichoplax adhaerens F.E. Schulze (Placozoa). Zoomorphology 98, 47–67. doi: 10.1007/BF00310320
Guidi, L., Eitel, M., Cesarini, E., Schierwater, B., and Balsamo, M. (2011). Ultrastructural analyses support different morphological lineages in the phylum Placozoa Grell, 1971. J. Morphol. 272, 371–378. doi: 10.1002/jmor.10922
Hedges, S. B., Blair, J. E., Venturi, M. L., and Shoe, J. L. (2004). A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 4:2. doi: 10.1186/1471-2148-4-2
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Jiménez-Guri, E., Philippe, H., Okamura, B., and Holland, P. W. H. (2007). Buddenbrockia is a cnidarian worm. Science 317, 116–118. doi: 10.1126/science.1142024
Kajitani, R., Toshimoto, K., Noguchi, H., Toyoda, A., Ogura, Y., Okuno, M., et al. (2014). Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24, 1384–1395. doi: 10.1101/gr.170720.113
Kamm, K., Osigus, H.-J., Stadler, P. F., DeSalle, R., and Schierwater, B. (2018). Trichoplax genomes reveal profound admixture and suggest stable wild populations without bisexual reproduction. Sci. Rep. 8:11168. doi: 10.1038/s41598-018-29400-y
Katoh, K., Misawa, K., Kuma, K.-I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Knaust, D. (2021). A microbialite with its entombed benthic community from the middle Triassic (Anisian-Ladinian) Muschelkalk group of Germany. Palaeontographica 320, 1–63. doi: 10.1127/pala/2021/0114
Komiya, Y., and Habas, R. (2008). Wnt signal transduction pathways. Organogenesis 4, 68–75. doi: 10.4161/org.4.2.5851
Kristensen, R. M. (1983). Loricifera, a new phylum with Aschelminthes characters from the meiobenthos. J. Zool. Syst. Evol. Res. 21, 163–180.
Kristensen, R. M., and Funch, P. (2000). Micrognathozoa: a new class with complicated jaws like those of Rotifera and Gnathostomulida. J. Morphol. 246, 1–49. doi: 10.1002/1097-4687(200010)246:1<1::AID-JMOR1>3.0.CO;2-D
Krumbach, T. (1907). Trichoplax, die umgewandelte Planula einer Hydromeduse. Zool. Anz. 31, 450–454.
Kuhl, W., and Kuhl, G. (1966). Untersuchungen über das Bewegungsverhalten von Trichoplax adhaerens F. E. Schulze (Zeittransformation: Zeitraffung). Z. Morphol. Okol. Tiere 56, 417–435. doi: 10.1007/BF00442291
Kumar, S., Suleski, M., Craig, J. M., Kasprowicz, A. E., Sanderford, M., Li, M., et al. (2022). TimeTree 5: an expanded resource for species divergence times. Mol. Biol. Evol. 39:msac174. doi: 10.1093/molbev/msac174
Laumer, C. E., Fernández, R., Lemer, S., Combosch, D., Kocot, K. M., Riesgo, A., et al. (2019). Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. Biol. Sci. 286:20190831. doi: 10.1098/rspb.2019.0831
Laumer, C. E., Gruber-Vodicka, H., Hadfield, M. G., Pearse, V. B., Riesgo, A., Marioni, J. C., et al. (2018). Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. elife 7:e36278. doi: 10.7554/eLife.36278
Lee, E. K., Cibrian-Jaramillo, A., Kolokotronis, S.-O., Katari, M. S., Stamatakis, A., Ott, M., et al. (2011). A functional phylogenomic view of the seed plants. PLoS Genet. 7:e1002411. doi: 10.1371/journal.pgen.1002411
Lin, H.-H., and Liao, Y.-C. (2016). Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci. Rep. 6:24175. doi: 10.1038/srep24175
Littlewood, D. T. J., Telford, M. J., Clough, K. A., and Rohde, K. (1998). Gnathostomulida—an enigmatic metazoan phylum from both morphological and molecular perspectives. Mol. Phylogenet. Evol. 9, 72–79. doi: 10.1006/mpev.1997.0448
Marques, A. C., and Collins, A. G. (2005). Cladistic analysis of Medusozoa and cnidarian evolution. Invertebr. Biol. 123, 23–42. doi: 10.1111/j.1744-7410.2004.tb00139.x
Miller, D. J., and Ball, E. E. (2005). Animal evolution: the enigmatic phylum Placozoa revisited. Curr. Biol. 15, R26–R28. doi: 10.1016/j.cub.2004.12.016
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Miranda, L. S., Hirano, Y. M., Mills, C. E., Falconer, A., Fenwick, D., Marques, A. C., et al. (2016). Systematics of stalked jellyfishes (Cnidaria: Staurozoa). PeerJ 4:e1951. doi: 10.7717/peerj.1951
Miyazawa, H., Osigus, H.-J., Rolfes, S., Kamm, K., Schierwater, B., and Nakano, H. (2021). Mitochondrial genome evolution of placozoans: gene rearrangements and repeat expansions. Genome Biol. Evol. 13:evaa213. doi: 10.1093/gbe/evaa213
Monticelli, F. S. (1895). Adelotacta Zoologica. 2. Treptoplax reptans Montic. Mittheilungen aus der Zoologischen Station zu Neapel, zugleich ein Repertorium für Mittelmeerkunde 12, 444–462.
Nesnidal, M. P., Helmkampf, M., Bruchhaus, I., El-Matbouli, M., and Hausdorf, B. (2013). Agent of whirling disease meets orphan worm: phylogenomic analyses firmly place Myxozoa in Cnidaria. PLoS One 8:e54576. doi: 10.1371/journal.pone.0054576
Neumann, J. S., DeSalle, R., Narechania, A., Schierwater, B., and Tessler, M. (2021a). Morphological characters can strongly influence early animal relationships inferred from phylogenomic data sets. Syst. Biol. 70, 360–375. doi: 10.1093/sysbio/syaa038
Neumann, J. S., Tessler, M., DeSalle, R., and Schierwater, B. (2021b). “Modern invertebrate systematics: the phylogenetics of early Metazoa” in Invertebrate Zoology: A Tree of Life Approach. eds. B. Schierwater and R. DeSalle (Boca Raton: CRC Press).
Nielsen, C. (2012). Animal Evolution: Interrelationships of the Living Phyla. Oxford Oxford University Press on Demand.
Nielsen, C. (2019). Early animal evolution: a morphologist’s view. R. Soc. Open Sci. 6:190638. doi: 10.1098/rsos.190638
Noll, F. C. (1890). “Über das Leben niederer Seetiere” in Bericht über die Senckenbergische naturforschende Gesellschaft in Frankfurt Am Main. 1890. ed. J. H. Rehn (Frankfurt am Main: Gebrüder Knauer).
Osigus, H.-J., Rolfes, S., Herzog, R., Kamm, K., and Schierwater, B. (2019). Polyplacotoma mediterranea is a new ramified placozoan species. Curr. Biol. 29, R148–R149. doi: 10.1016/j.cub.2019.01.068
Paknia, O., and Schierwater, B. (2015). Global habitat suitability and ecological niche separation in the phylum Placozoa. PLoS One 10:e0140162. doi: 10.1371/journal.pone.0140162
Pandey, A., and Braun, E. L. (2020). Phylogenetic analyses of sites in different protein structural environments result in distinct placements of the metazoan root. Biology 9:64. doi: 10.3390/biology9040064
Paradis, E. (2013). Molecular dating of phylogenies by likelihood methods: a comparison of models and a new information criterion. Mol. Phylogenet. Evol. 67, 436–444. doi: 10.1016/j.ympev.2013.02.008
Paradis, E., and Schliep, K. (2019). Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Pearse, V. B., and Voigt, O. (2007). Field biology of placozoans (Trichoplax): distribution, diversity, biotic interactions. Integr. Comp. Biol. 47, 677–692. doi: 10.1093/icb/icm015
Peterson, K. J., and Butterfield, N. J. (2005). Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc. Natl. Acad. Sci. U. S. A. 102, 9547–9552. doi: 10.1073/pnas.0503660102
Peterson, K. J., and Eernisse, D. J. (2001). Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol. Dev. 3, 170–205. doi: 10.1046/j.1525-142x.2001.003003170.x
Pett, W., Adamski, M., Adamska, M., Francis, W. R., Eitel, M., Pisani, D., et al. (2019). The role of homology and orthology in the phylogenomic analysis of metazoan gene content. Mol. Biol. Evol. 36, 643–649. doi: 10.1093/molbev/msz013
Philippe, H., Derelle, R., Lopez, P., Pick, K., Borchiellini, C., Boury-Esnault, N., et al. (2009). Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712. doi: 10.1016/j.cub.2009.02.052
Podar, M., Haddock, S. H., Sogin, M. L., and Harbison, G. R. (2001). A molecular phylogenetic framework for the phylum Ctenophora using 18S rRNA genes. Mol. Phylogenet. Evol. 21, 218–230. doi: 10.1006/mpev.2001.1036
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., and Korobeynikov, A. (2020). Using SPAdes De novo assembler. Curr. Protoc. Bioinformatics 70:e102. doi: 10.1002/cpbi.102
R Core Team (2020). R: a language and environment for statistical computing. Available at: https://www.R-project.org/ (Accessed 01 October, 2020).
Rahnenfuhrer, A. (2022). TopGo: Enrichment Analysis for Gene Ontology R package version 2.44.0. Bioconductor 3.15. doi: 10.18129/B9.bioc.topGO
Revell, L. J. (2012). Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Romanova, D. Y., Varoqueaux, F., Daraspe, J., Nikitin, M. A., Eitel, M., Fasshauer, D., et al. (2021). Hidden cell diversity in Placozoa: ultrastructural insights from Hoilungia hongkongensis. Cell Tissue Res. 385, 623–637. doi: 10.1007/s00441-021-03459-y
Ruiz-Trillo, I., Inagaki, Y., Davis, L. A., Sperstad, S., Landfald, B., and Roger, A. J. (2004). Capsaspora owczarzaki is an independent opisthokont lineage. Curr. Biol. 14, R946–R947. doi: 10.1016/j.cub.2004.10.037
Rutledge, L. Y., Garroway, C. J., Loveless, K. M., and Patterson, B. R. (2010). Genetic differentiation of eastern wolves in Algonquin Park despite bridging gene flow between coyotes and grey wolves. Heredity 105, 520–531. doi: 10.1038/hdy.2010.6
Ryan, J. F., Pang, K., Schnitzler, C. E., Nguyen, A.-D., Moreland, R. T., Simmons, D. K., et al. (2013). The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342:1242592. doi: 10.1126/science.1242592
Schierwater, B. (2005). My favorite animal, Trichoplax adhaerens. BioEssays 27, 1294–1302. doi: 10.1002/bies.20320
Schierwater, B., Eitel, M., Jakob, W., Osigus, H.-J., Hadrys, H., Dellaporta, S. L., et al. (2009). Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 7:e1000020:e20. doi: 10.1371/journal.pbio.1000020
Schierwater, B., Eitel, M., Osigus, H.-J., von der Chevallerie, K., Bergmann, T., Hadrys, H., et al. (2010). “Trichoplax and Placozoa: one of the crucial keys to understanding metazoan evolution” in Key Transitions in Animal Evolution. eds. R. Desalle and B. Schierwater (Rawalpindi: Science Publishers).
Schierwater, B., Osigus, H.-J., Bergmann, T., Blackstone, N. W., Hadrys, H., Hauslage, J., et al. (2021a). The enigmatic Placozoa part 1: exploring evolutionary controversies and poor ecological knowledge. BioEssays 43:e2100080. doi: 10.1002/bies.202100080
Schierwater, B., Osigus, H., Bergmann, T., Blackstone, N. W., Hadrys, H., Hauslage, J., et al. (2021b). The enigmatic Placozoa part 2: exploring evolutionary controversies and promising questions on earth and in space. BioEssays 43:e2100083. doi: 10.1002/bies.202100083
Sebé-Pedrós, A., Burkhardt, P., Sánchez-Pons, N., Fairclough, S. R., Lang, B. F., King, N., et al. (2013). Insights into the origin of metazoan filopodia and microvilli. Mol. Biol. Evol. 30, 2013–2023. doi: 10.1093/molbev/mst110
Signorovitch, A. Y., Buss, L. W., and Dellaporta, S. L. (2007). Comparative genomics of large mitochondria in placozoans. PLoS Genet. 3:e13. doi: 10.1371/journal.pgen.0030013
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Simion, P., Bekkouche, N., Jager, M., Quéinnec, E., and Manuel, M. (2015). Exploring the potential of small RNA subunit and ITS sequences for resolving phylogenetic relationships within the phylum Ctenophora. Zoology 118, 102–114. doi: 10.1016/j.zool.2014.06.004
Simion, P., Philippe, H., Baurain, D., Jager, M., Richter, D. J., Di Franco, A., et al. (2017). A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 27, 958–967. doi: 10.1016/j.cub.2017.02.031
Smit, A. F. A., and Hubley, R. (2015). RepeatModeler Open-1.0. Available at: http://www.repeatmasker.org/ (Accessed September 08, 2018).
Smith, C. L., Varoqueaux, F., Kittelmann, M., Azzam, R. N., Cooper, B., Winters, C. A., et al. (2014). Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 24, 1565–1572. doi: 10.1016/j.cub.2014.05.046
Srivastava, M., Begovic, E., Chapman, J., Putnam, N. H., Hellsten, U., Kawashima, T., et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454, 955–960. doi: 10.1038/nature07191
Suga, H., Chen, Z., de Mendoza, A., Sebé-Pedrós, A., Brown, M. W., Kramer, E., et al. (2013). The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 4:2325. doi: 10.1038/ncomms3325
Syed, T., and Schierwater, B. (2002a). Trichoplax adhaerens: discovered as a missing link, forgotten as a hydrozoan, re-discovered as a key to metazoan evolution. Vie Milieu Paris 52, 177–187.
Syed, T., and Schierwater, B. (2002b). The evolution of the Placozoa: a new morphological model. Senckenb. Lethaea 82, 315–324. doi: 10.1007/BF03043791
Tessler, M., Galen, S. C., DeSalle, R., and Schierwater, B. (2022). Let’s end taxonomic blank slates with molecular morphology. Front. Ecol. Evol. 10:1016412.
UniProt Consortium (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi: 10.1093/nar/gkaa1100
Voigt, O., Collins, A. G., Pearse, V. B., Pearse, J. S., Ender, A., Hadrys, H., et al. (2004). Placozoa -- no longer a phylum of one. Curr. Biol. 14, R944–R945. doi: 10.1016/j.cub.2004.10.036
Wallberg, A., Thollesson, M., Farris, J. S., and Jondelius, U. (2004). The phylogenetic position of the comb jellies (Ctenophora) and the importance of taxonomic sampling. Cladistics 20, 558–578. doi: 10.1111/j.1096-0031.2004.00041.x
Wheeldon, T. J., and Patterson, B. R. (2012). Genetic and morphological differentiation of wolves (Canis lupus) and coyotes (Canis latrans) in northeastern Ontario. Can. J. Zool. 90, 1221–1230. doi: 10.1139/z2012-090
Whelan, N. V., Kocot, K. M., Moroz, L. L., and Halanych, K. M. (2015). Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl. Acad. Sci. U. S. A. 112, 5773–5778. doi: 10.1073/pnas.1503453112
Whelan, N. V., Kocot, K. M., Moroz, T. P., Mukherjee, K., Williams, P., Paulay, G., et al. (2017). Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737–1746. doi: 10.1038/s41559-017-0331-3
Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. Berlin: Springer Science & Business Media.
WoRMS Editorial Board (2021). World Register of Marine Species. Available at: https://www.marinespecies.org at VLIZ. (Accessed October 27, 2022).
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. (2017). Ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Keywords: molecular morphology, higher taxonomy, Metazoa, morphology, phylogenomics, Placozoa
Citation: Tessler M, Neumann JS, Kamm K, Osigus H-J, Eshel G, Narechania A, Burns JA, DeSalle R and Schierwater B (2022) Phylogenomics and the first higher taxonomy of Placozoa, an ancient and enigmatic animal phylum. Front. Ecol. Evol. 10:1016357. doi: 10.3389/fevo.2022.1016357
Edited by:
Rodney L. Honeycutt, Pepperdine University, United StatesReviewed by:
Christopher A. Emerling, Reedley College, United StatesDaria Romanova, Institute of Higher Nervous Activity and Neurophysiology (RAS), Russia
Copyright © 2022 Tessler, Neumann, Kamm, Osigus, Eshel, Narechania, Burns, DeSalle and Schierwater. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Tessler, mtessler@sfc.edu; Johannes S. Neumann, jneumann@amnh.org; Bernd Schierwater, bernd.schierwater@tiho-hannover.de
†These authors share first authorship
 Michael Tessler
Michael Tessler Johannes S. Neumann
Johannes S. Neumann Kai Kamm
Kai Kamm Hans-Jürgen Osigus
Hans-Jürgen Osigus Gil Eshel
Gil Eshel Apurva Narechania2
Apurva Narechania2  John A. Burns
John A. Burns Rob DeSalle
Rob DeSalle Bernd Schierwater
Bernd Schierwater