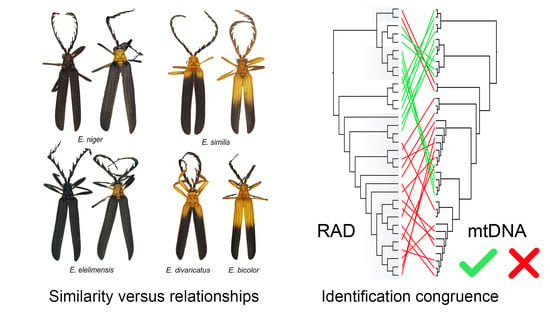
Genomic and Mitochondrial Data Identify Different Species Boundaries in Aposematically Polymorphic Eniclases Net-Winged Beetles (Coleoptera: Lycidae)
Abstract
:1. Introduction
2. Methods
2.1. Material and Laboratory Procedures
2.2. Mitochondrial DNA Data Sampling and Phylogenetic Analyses
2.3. Next-RAD Sampling, Filtering and SNP Calling
2.4. RAD Tree Processing
2.5. Morphology
3. Results
3.1. Morphology
3.2. MtDNA Analysis
3.3. Next-RAD Analysis and Phylogeny
4. Discussion
4.1. Species Delimitation: Congruence and Conflicts
4.2. Taxonomy of Eniclases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeSalle, R.; Egan, M.G.; Siddall, M. The unholy trinity: Taxonomy, species delimitation and DNA barcoding. Phil. Trans. R. Soc. Biol. Sci. 2005, 360, 1905–1916. [Google Scholar] [CrossRef] [PubMed]
- Larson, W.A.; Seeb, L.W.; Everett, M.V.; Waples, R.K.; Templin, W.D.; Seeb, J.E. Genotyping by sequencing resolves shallow population structure to inform conservation of Chinook salmon (Oncorhynchus tshawytscha). Evol. Appl. 2014, 7, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Nater, A.; Mattle-Greminger, M.P.; Nurcahyo, A.; Nowak, M.G.; de Manuel, M.; Desai, T.; Groves, C.; Pybus, M.; Sonay, T.B.; Roos, C.; et al. Morphometric, behavioral, and genomic evidence for a new orangutan species. Curr. Biol. 2017, 27, 3487–3498. [Google Scholar] [CrossRef] [PubMed]
- Abdelkrim, J.; Aznar-Cormano, L.; Buge, B.; Fedosov, A.; Kantor, Y.; Zaharias, P.; Puillandre, N. Delimiting species of marine gastropods (Turridae, Conoidea) using RAD sequencing in an integrative taxonomy framework. Mol. Ecol. 2018, 27, 4591–4611. [Google Scholar] [CrossRef] [PubMed]
- Riedel, A.; Sagata, K.; Surbakti, S.; Tanzler, R.; Balke, M. One hundred and one new species of Trigonopterus weevils from New Guinea. ZooKeys 2013, 280, 1–150. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, D.; Monaghan, M.T.; Vogler, A.P. DNA-based taxonomy for associating adults and larvae in multi-species assemblages of chafers (Coleoptera: Scarabaeidae). Mol. Phyl. Evol. 2007, 44, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, D.; Fujisawa, T.; Krammer, H.-J.; Eberle, J.; Fabrizi, S.; Vogler, A.P. Rarity and incomplete sampling in DNA-based species delimitation. Syst. Biol. 2016, 65, 478–494. [Google Scholar] [CrossRef] [PubMed]
- Riedel, A.; Tanzler, R.; Pons, J.; Suhardjono, Y.R.; Balke, M. Large-scale molecular phylogeny of Cryptorhynchinae (Coleoptera, Curculionidae) from multiple genes suggests American origin and later Australian radiation. Syst. Entomol. 2016, 41, 492–503. [Google Scholar] [CrossRef]
- Li, Y.; Gunter, N.; Hong, P.; Bocak, L. DNA-based species delimitation separates highly divergent populations within morphologically coherent clades of poorly dispersing beetles. Zool. J. Linn. Soc. 2015, 175, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Morinière, J.; Cancian de Araujo, B.; Lam, A.W.; Hausmann, A.; Balke, M.; Schmidt, S.; Hendrich, L.; Doczkal, D.; Fartmann, B.; Arvidsson, S.; et al. Species identification in malaise trap samples by DNA barcoding based on NGS technologies and a scoring matrix. PLoS ONE 2016, 11, e0155497. [Google Scholar] [CrossRef]
- Cruaud, A.; Gautier, M.; Galan, M.; Foucaud, J.; Saune, L.; Genson, G.; Dubois, E.; Nidelet, S.; Deuve, T.; Rasplus, J.-Y. Empirical assessment of RAD sequencing for interspecific phylogeny. Mol. Biol. Evol. 2014, 31, 1272–1274. [Google Scholar] [CrossRef] [PubMed]
- Bray, T.C.; Bocak, L. Slowly dispersing neotenic beetles can speciate on a penny coin and generate space-limited diversity in the tropical mountains. Sci. Rep. 2016, 6, 33579. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Sota, T. Divergent host use among cryptic species in the fungivorous ciid beetle Octotemnus laminifrons (Motschulsky, 1860), with descriptions of three new species from Japan. Syst. Entomol. 2019, 44, 179–191. [Google Scholar] [CrossRef]
- Herrera, S.; Shank, T.M. RAD sequencing enables unprecedented phylogenetic resolution and objective species delimitation in recalcitrant divergent taxa. Mol. Phyl. Evol. 2016, 100, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sklenarova, K.; Kubecek, V.; Bocak, L. Subtribal classification of Metriorrhynchini (Insecta: Coleoptera: Lycidae): An integrative approach using molecular phylogeny and morphology of adults and larvae. Arthr. Syst. Phyl. 2014, 72, 37–54. [Google Scholar]
- Bocak, L.; Bocakova, M. Revision of the genus Eniclases Waterhouse, 1879 (Coleoptera, Lycidae, Metriorrhynchinae). Mitt. Münch. Entomol. Ges. 1991, 81, 203–226. [Google Scholar]
- Waterhouse, C.O. Illustration of the Typical Specimens of Coleoptera in the Collection of the British Museum. In Part I.—Lycidae; British Museum: London, UK, 1879. [Google Scholar]
- Pic, M. Contribution à l’étude des Lycides. L’Echange 1921, 406, 9–12. [Google Scholar]
- Kleine, R. Coleoptera—Lycidae. Nova Guin. 1926, 15, 91–195. [Google Scholar]
- Bocek, M.; Bocak, L. Species limits in polymorphic mimetic Eniclases net-winged beetles from New Guinean mountains (Coleoptera: Lycidae). Zookeys 2016, 593, 15–35. [Google Scholar]
- Bocek, M.; Adamkova, K. New species of trichaline net-winged beetles, with remarks on the phylogenetic position and distribution of Schizotrichalus (Coleoptera: Lycidae: Metriorrhynchinae). Zootaxa 2019, 4623, 341–350. [Google Scholar] [CrossRef]
- Bocek, M.; Kusy, D.; Motyka, M.; Bocak, L. Persistence of multiple patterns and intraspecific polymorphism in multi-species Müllerian communities of net-winged beetles. Front. Zool. (accepted).
- Sklenarova, K.; Chesters, D.; Bocak, L. Phylogeography of poorly dispersing net-winged beetles: A role of drifting India in the origin of Afrotropical and Oriental fauna. PLoS ONE 2013, 8, e67957. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinform. 2008, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Meth. 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ekblom, R.; Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 2011, 107, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Eaton, D.A.R. PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 2014, 30, 1844–1849. [Google Scholar] [CrossRef]
- Eaton, D.A.R.; Overcast, I. iPYRAD: Interactive Assembly and Analysis of RADseq Data Sets. 2016. Available online: https://ipyrad.readthedocs.io/ (accessed on 20 April 2019).
- Linsley, E.G.; Eisner, T.; Klots, A.B. Mimetic assemblages of sibling species of lycid beetles. Evolution 1961, 15, 15–29. [Google Scholar] [CrossRef]
- Eisner, T.; Kafatos, F.C.; Linsley, E.G. Lycid predation by mimetic adult Cerambycidae (Coleoptera). Evolution 1962, 16, 316–324. [Google Scholar] [CrossRef]
- Moore, B.P.; Brown, W.V. Identification of warning odour components, bitter principles and antifeedants in an aposematic beetle: Metriorrhynchus rhipidius (Coleoptera: Lycidae). Ins. Biochem. 1981, 11, 493–499. [Google Scholar] [CrossRef]
- Eisner, T.; Schroeder, F.C.; Snyder, N.; Grant, J.B.; Aneshansley, D.J.; Utterback, D.; Meinwald, J.; Eisner, M. Defensive chemistry of lycid beetles and of mimetic cerambycid beetles that feed on them. Chemoecology 2008, 18, 109–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocak, L.; Yagi, T. Evolution of mimicry patterns in Metriorrhynchus (Coleoptera, Lycidae): E history of dispersal and speciation in Southeast Asia. Evolution 2010, 64, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Motyka, M.; Kampova, L.; Bocak, L. Phylogeny and evolution of Müllerian mimicry in aposematic Dilophotes: Evidence for advergence and size-constraints in evolution of mimetic sexual dimorphism. Sci. Rep. 2018, 8, 3744. [Google Scholar] [CrossRef] [PubMed]
- Mallet, J. A species definition for the modern synthesis. Trends Ecol. Evol. 1995, 10, 294–299. [Google Scholar] [CrossRef]
- Dupuis, J.R.; Roe, A.D.; Sperling, F.A.H. Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol. Ecol. 2012, 21, 4422–4436. [Google Scholar] [CrossRef] [PubMed]
- Leache, A.; J Oaks, J.R. The Utility of Single Nucleotide Polymorphism (SNP) Data in Phylogenetics. Ann. Rev. Ecol. Evol. Syst. 2017, 48, 69–84. [Google Scholar] [CrossRef]
- Ivanov, V.; Lee, K.M.; Mutanen, M. Mitonuclear discordance in wolf spiders: Genomic evidence for species integrity and introgression. Mol. Ecol. 2018, 27, 1681–1695. [Google Scholar] [CrossRef]
- Tocco, C.; Dacke, M.; Byrne, M. Eye and wing structure closely reflects the visual ecology of dung beetles. J. Comp. Physiol. A Neuroethol. Sens. Neur. Behav. Physiol. 2019, 205, 211–221. [Google Scholar] [CrossRef]
- Mayr, E. Systematics and the Origin of Species, from the Viewpoint of a Zoologist; Harvard University Press: Cambridge, MA, USA, 1942. [Google Scholar]
- Cracraft, J. Species concepts and speciation analysis. Curr. Ornith. 1983, 1, 159–187. [Google Scholar]
- Coyne, J.A.; Orr, H.A. Speciation; Sinauer Associates: Sunderland, MA, USA, 2004. [Google Scholar]
- Mallet, J. Hybridization, ecological races and the nature of species: Empirical evidence for the ease of speciation. Phil. Trans. R. Soc. Biol. Sci. 2008, 363, 2971–2986. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic diversity and conservation units: dealing with the species-population continuum in the age of genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Rosser, N.; Freitas, A.V.L.; Huertas, B.; Joron, M.; Lamas, G.; Mérot, C.; Simpson, F.; Willmott, K.R.; Mallet, J.; Dasmahapatra, K.K. Cryptic speciation associated with geographic and ecological divergence in two Amazonian Heliconius butterflies. Zool. J. Linn. Soc. 2019, 186, 233–249. [Google Scholar] [CrossRef]
- Linck, E.; Epperly, K.; Van Els, P.; Spellman, G.M.; Bryson, R.W., Jr.; McCormack, J.E.; Canales-Del-Castillo, R.; Klicka, J. Dense geographic and genomic sampling reveals paraphyly and a cryptic lineage in a classic sibling species complex. Syst. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Elias, M.; Hill, R.I.; Willmott, K.R.; Dasmahapatra, K.K.; Brower, A.V.Z.; Mallet, J.; Jiggins, C.D. Limited performance of DNA barcoding in a diverse community of tropical butterflies. Proc. R. Soc. Biol. Sci. 2007, 274, 2881–2889. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Eniclases Identification Genomic |
mtDNA and morph. | Voucher Number (UPOL+) |
Locality (All New Guinea; See Figure 2A) |
|
|---|---|---|---|---|
| E. pseudoapertus | E. pseudoapertus | BM0080 | Arfak Mts., Maibri, 1570 m | |
| E. pseudoluteolus | E. pseudoluteolus | BM0084 | Arfak Mts., Maibri, 1570 m | |
| E. divaricatus | E. divaricatus | BM0057 | Elelim, 580 m | |
| E. divaricatus | E. divaricatus | BM0001, 02, 09, 15–17 | Sentani, 275 m | |
| E. apertus | E. apertus | BM0038 | Sentani, 360 m | |
| E. apertus | E. apertus | BM0018 | Sentani, 275 m | |
| E. niger | P | E. niger | BM0087, 89 | Bokondini, 1287 m |
| E. niger | P | E. niger | BM0058–61 | Elelim, 580 m |
| E. similis | P | E. similis | BM0024 | Sentani, 275 m |
| E. similis | P | E. similis | BM0037 | Sentani, 360 m |
| E. similis | P | E. similis | BM0003–04, 19–23, 11, 13–14 | Sentani, 275 m |
| E. infuscatus | E. infuscatus | BM0050 | Elelim, 580 m | |
| E. infuscatus | E. infuscatus | BM0062 | Elelim, 650 m | |
| E. bicolor | E. bicolor | BM0045–47 | Elelim, 580 m | |
| E. tikapurensis | Eniclases sp. A | BM0093 | Bokondini, 1750–1900 m | |
| E. tikapurensis | Eniclases sp. A | BM0096–97 | Bokondini, 2100 m | |
| E. tikapurensis | E. tikapurensis | BM0039 | Yiwika, 2100 m | |
| E. tikapurensis | E. tikapurensis | BM0040–44 | Tikapura, 2170 m | |
| Eniclases sp. B | P | E. variabilis | BM0008, 12 | Sentani, 275 m |
| E. brancuccii | E. brancuccii | BM0005–07, 10 | Sentani, 275 m | |
| E. bokondinensis | E. bokondinensis | BM0092, 94–95 | Bokondini, 1750–1900 m | |
| E. variabilis | P | E. variabilis | BM0048–49, 54–55 | Elelim, 580 m |
| E. elelimensis | P | E. elelimensis | BM0051–52, 56 | Elelim, 580 m |
| E. elelimensis | P | E. variabilis | BM0053 | Elelim, 580 m |
| E. elelimensis | P | E. variabilis | BM0027–29, 35–36 | Bokondini, 1250–1300 m |
| E. elelimensis | P | E. variabilis | BM0086, 88, 90–91 | Bokondini, 1287 m |
| E. elelimensis | P | E. variabilis | BM0063–67 | Dombomi, 1150 m |
| E. elelimensis | P | E. variabilis | BM0025–26, 3–32, 34 | Bokondini1250–1300 m |
| Species | Body Length |
Width Humeri |
Pronotum Length |
Width | EDiam/EDist Male |
|---|---|---|---|---|---|
| E. pseudoapertus | 6.3 | 1.6 | 0.75 | 1.2 | 1.4 |
| E. divaricatus | 6.8–9.7 | 2.1–2.3 | 1.2–1.3 | 1.7–1.7 | 0.92–0.96 |
| E. pseudoluteolus | 9.3 | 2.3 | 1.15 | 1.6 | 0.9 |
| E. apertus | 5.7–8.4 | 1.34–1.7 | 0.9 | 1.25 | 1.15–1.17 |
| E. tikapurensis | 9.5–11.1 | 2.0–2.5 | 1.1–1.3 | 1.4–1.7 | 1.11–1.40 |
| E. bicolor | 10.3 | 2.4 | 1.4 | 1.7 | n.a. |
| E. infuscatus | 12.1 | 2.5 | 1.25 | 1.6 | n.a. |
| E. brancuccii | 7.6–8.0 | 1.8–1.9 | 1.0–1.1 | 1.5–1.8 | 1.0 |
| E. bokondinensis | 9.2 | 2.05 | 1.0 | 1.35 | n.a. |
| E. elelimensis | 6.9–8.1 | 1.5–1.9 | 0.9–1.1 | 1.3–1.4 | n.a. |
| E. variabilis | 6.6–8.2 | 1.6–2.0 | 0.1–1.1 | 1.1–1.35 | 0.83–0.95 |
| E. niger | 9.2–11.6 | 2.2–2.8 | 1.3–1.6 | 1.3–1.6 | 1.17–1.28 |
| E. similis | 7.5–9.7 | 1.9–2.3 | 1.1–1.4 | 1.8 | 1.02–1.1 |
| div | sim | nig | bra | var/B | var | elel | bok | ape | tik | tik/A | bic | inf | psl | psap | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM0001 E. divaricatus |
- | ||||||||||||||
| BM0003 E. similis |
12.08 | ||||||||||||||
| BM0033 E. niger |
12.35 | 0.45 | - | ||||||||||||
| BM0005 E. brancuccii |
12.53 | 8.63 | 8.17 | - | |||||||||||
| BM0008 E. sp. B/variabilis |
12.72 | 8.63 | 8.17 | 3.91 | - | ||||||||||
| BM0054 E. variabilis |
12.72 | 8.54 | 8.08 | 4.27 | 0.45 | - | |||||||||
| BM0051 E. elelimensis |
12.72 | 8.08 | 7.63 | 3.81 | 1.54 | 1.63 | - | ||||||||
| BM0092 E. bokondinensis | 13.08 | 8.17 | 7.72 | 4 | 1.73 | 1.82 | 0.73 | - | |||||||
| BM0018 E. apertus |
13.17 | 10.26 | 10.17 | 10.08 | 10.35 | 10.54 | 9.72 | 10.08 | - | ||||||
| BM0039 E. tikapurensis |
13.17 | 10.08 | 9.99 | 9.81 | 9.99 | 10.17 | 8.99 | 9.45 | 6.81 | - | |||||
| BM0093 E. tikapur./sp.A |
13.62 | 9.81 | 9.72 | 9.9 | 9.9 | 9.99 | 9.08 | 9.54 | 7.27 | 1.18 | - | ||||
| BM0045 E. bicolor |
12.90 | 10.54 | 10.45 | 9.63 | 9.81 | 9.9 | 8.99 | 9.08 | 8.72 | 6.45 | 6.27 | - | |||
| BM0050 E. infuscatus |
12.72 | 9.99 | 10.08 | 9.45 | 9.54 | 9.54 | 8.54 | 8.63 | 8.72 | 6.18 | 6.18 | 1.09 | - | ||
| BM0084 E. pseudoluteolus |
12.99 | 9.45 | 9.26 | 10.26 | 10.9 | 10.81 | 9.81 | 10.26 | 12.08 | 10.81 | 11.17 | 11.81 | 11.26 | - | |
| BM0080 E. pseudoapertus |
13.26 | 10.9 | 10.9 | 10.9 | 11.44 | 11.35 | 10.99 | 11.44 | 11.44 | 11.35 | 11.35 | 11.08 | 10.63 | 11.53 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bocek, M.; Motyka, M.; Kusy, D.; Bocak, L. Genomic and Mitochondrial Data Identify Different Species Boundaries in Aposematically Polymorphic Eniclases Net-Winged Beetles (Coleoptera: Lycidae). Insects 2019, 10, 295. https://doi.org/10.3390/insects10090295
Bocek M, Motyka M, Kusy D, Bocak L. Genomic and Mitochondrial Data Identify Different Species Boundaries in Aposematically Polymorphic Eniclases Net-Winged Beetles (Coleoptera: Lycidae). Insects. 2019; 10(9):295. https://doi.org/10.3390/insects10090295
Chicago/Turabian StyleBocek, Matej, Michal Motyka, Dominik Kusy, and Ladislav Bocak. 2019. "Genomic and Mitochondrial Data Identify Different Species Boundaries in Aposematically Polymorphic Eniclases Net-Winged Beetles (Coleoptera: Lycidae)" Insects 10, no. 9: 295. https://doi.org/10.3390/insects10090295