Abstract
Ring species provide particularly clear demonstrations of how one species can gradually evolve into two, but are rare in nature1,2,3. In the greenish warbler (Phylloscopus trochiloides) species complex, a ring of populations wraps around Tibet. Two reproductively isolated forms co-exist in central Siberia, with a gradient of genetic and phenotypic characteristics through the southern chain of populations connecting them4,5,6. Previous genetic evidence has proven inconclusive, however, regarding whether species divergence took place in the face of continuous gene flow and whether hybridization between the terminal forms of the ring ever occurred7,8,9. Here we use genome-wide analyses to show that, although spatial patterns of genetic variation are currently mostly as expected of a ring species, historical breaks in gene flow have existed at more than one location around the ring, and the two Siberian forms have occasionally interbred. Substantial periods of geographical isolation occurred not only in the north but also in the western Himalayas, where there is now an extensive hybrid zone between genetically divergent forms. Limited asymmetric introgression has occurred directly between the Siberian forms, although it has not caused a blending of those forms, suggesting selection against introgressed genes in the novel genetic background. Levels of reproductive isolation and genetic introgression are consistent with levels of phenotypic divergence around the ring, with phenotypic similarity and extensive interbreeding across the southwestern contact zone and strong phenotypic divergence and nearly complete reproductive isolation across the northern contact zone. These results cast doubt on the hypothesis that the greenish warbler should be viewed as a rare example of speciation by distance6, but demonstrate that the greenish warbler displays a continuum from slightly divergent neighbouring populations to almost fully reproductively isolated species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Data deposits
Raw Illumina sequencing data have been deposited in the Sequence Read Archive (SRA) under accession numbers SRX472921 and SRX473141. The list of contigs and information concerning the physical location of the SNP markers used in this study, using the zebra finch genome assembly as reference, have been deposited in the Dryad repository (http://doi.org/10.5061/dryad.6kn93).
References
Irwin, D. E., Irwin, J. H. & Price, T. D. Ring species as bridges between microevolution and speciation. Genetica 112–113, 223–243 (2001)
Moritz, C., Schneider, C. J. & Wake, D. B. Evolutionary relationships within the Ensatina eschscholtzii complex confirm the ring species interpretation. Syst. Biol. 41, 273–291 (1992)
Cacho, N. I. & Baum, D. A. The Caribbean slipper spurge Euphorbia tithymaloides: the first example of a ring species in plants. Proc. R. Soc. B 279, 3377–3383 (2012)
Irwin, D. E., Bensch, S. & Price, T. D. Speciation in a ring. Nature 409, 333–337 (2001)
Irwin, D. E., Thimgan, M. P. & Irwin, J. H. Call divergence is correlated with geographic and genetic distance in greenish warblers (Phylloscopus trochiloides): a strong role for stochasticity in signal evolution? J. Evol. Biol. 21, 435–448 (2008)
Irwin, D. E., Bensch, S., Irwin, J. H. & Price, T. D. Speciation by distance in a ring species. Science 307, 414–416 (2005)
Wake, D. B. Speciation in the round. Nature 409, 299–300 (2001)
Coyne, J. A. & Orr, H. A. Speciation (Sinauer Associates, 2004)
Kovylov, N. S., Marova, I. M. & Ivanitskii, V. V. Variation of song and plumage in the western (Phylloscopus trochiloides viridanus) and eastern (Phylloscopus trochiloides plumbeitarsus) forms of the greenish warbler in a sympatry zone: is the hypothesis of ring speciation true? Biol. Bull. 39, 729–740 (2012)
Mayr, E. Systematics and the Origin of Species (Columbia Univ. Press, 1942)
Price, T. D. Speciation in Birds (Roberts and Co., 2008)
Endler, J. A. Geographic Variation, Speciation, and Clines (Princeton Univ. Press, 1977)
Mayr, E. Animal Species and Evolution (Belknap, 1963)
Weir, J. T. & Price, T. D. Limits to speciation inferred from times to secondary sympatry and ages of hybridizing species along a latitudinal gradient. Am. Nat. 177, 462–469 (2011)
Ticehurst, C. B. A Systematic Review of the Genus Phylloscopus (Trustees of the British Museum, London, 1938)
Irwin, D. E. in Creating Consilience: Integrating the Sciences and the Humanities (eds Slingerland, E. & Collard, M.) 163–178 (Oxford Univ. Press, 2012)
Novembre, J. & Stephens, M. Interpreting principal component analyses of spatial population genetic variation. Nature Genet. 40, 646–649 (2008)
Martins, A. B., Aguiar, M. A. M. & Bar-Yam, Y. Evolution and stability of ring species. Proc. Natl Acad. Sci. USA 110, 5080–5084 (2013)
Irwin, D. E. Local adaptation along smooth ecological gradients causes phylogeographic breaks and phenotypic clustering. Am. Nat. 180, 35–49 (2012)
Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, e19379 (2011)
Barton, N. H. & Hewitt, G. M. Adaptation, speciation and hybrid zones. Nature 341, 497–503 (1989)
Rheindt, F. E. & Edwards, S. V. Genetic introgression: an integral but neglected component of speciation in birds. Auk 128, 620–632 (2011)
Irwin, D. E. & Irwin, J. H. in Birds of Two Worlds: The Ecology and Evolution of Migration (eds Greenberg, R. & Marra, P. P.) 27–40 (John Hopkins Univ. Press, 2005)
Price, T. D. & Bouvier, M. M. The evolution of F1 postzygotic incompatibilities in birds. Evolution 56, 2083–2089 (2002)
McCarthy, E. M. Handbook of Avian Hybrids of the World (Oxford Univ. Press, 2006)
Grant, P. R. & Grant, B. R. Hybridization of bird species. Science 256, 193–197 (1992)
Dobzhansky, T. in A Century of Darwin (ed. Barnett, S. A.) 19–55 (Heinemann, 1958)
Warren, W. C. et al. The genome of a songbird. Nature 464, 757–762 (2010)
Peakall, R. & Smouse, P. E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research — an update. Bioinformatics 28, 2537–2539 (2012)
Pritchard, J. K., Stephens, J. K. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000)
Drummond, A. J. et al. Geneious v5. 4 http://www.geneious.com/ (2011)
Evanno, G., Regnaut, S. J. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005)
Earl, D. A. & von Holdt, B. M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (2012)
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007)
Acknowledgements
This research was supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme (project no. 273773) and an NSERC discovery grant (no. 311931). Original collections were made with a grant from the US NSF. We thank Z. Benowitz-Fredericks, J. Gibson, S. Gross, J. Irwin, G. Kelberg, A. Knorre, K. Marchetti and B. Sheldon for assistance in the field. For additional samples we thank P. Alström, K. Marchetti, U. Olsson, A. Richman and J. Tiainen. We thank J. Irwin and L. Rieseberg for comments and discussion and A. Kuzmin for assistance during library preparation.
Author information
Authors and Affiliations
Contributions
M.A. and D.E.I. conceived the genomic study. M.A. designed the genomic analysis, and collected and analysed the sequence data. D.E.I., E.S.C.S. and T.D.P. conducted fieldwork. D.E.I. conducted the cline analysis in the southwestern contact zone. All authors contributed to the writing of the manuscript and have read and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
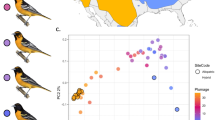
Extended Data Figure 1 Sampled sites, geographical variation in SNPs and genetic admixtures.
a, Geographical distribution of the sites sampled in this study (see also Extended Data Table 1). Colours correspond to the six subspecies of greenish warblers described in ref. 15 (P.t. trochiloides, yellow; ludlowi, green; obscuratus, orange; nitidus, violet; viridanus, blue; plumbeitarsus, red). Greenish warblers coexisting in central Siberia were classified as viridanus or plumbeitarsus according to their mtDNA, which for males was perfectly concordant with their song4. b, Geographic variation in 2,334 SNP markers across 95 greenish warblers as summarized by PCA. Individual birds are depicted as diamonds and colours represent their geographical origin as shown in a. c, Plot of genetic admixture proportions (Q) according to the five genetic clusters inferred by the software STRUCTURE30. Location names are given in the plot. We also indicate whether a particular individual displays a western (W) or eastern (E) mtDNA haplotype according to ref. 4.
Extended Data Figure 2 Genetic differentiation based on SNP markers increases with corrected geographical distance around the ring.
See ref. 6 for geographical distance; Mantel’s r = 0.71, P < 0.0001. Pair-wise FST values were calculated among those locations where at least 4 individuals were genotyped (N = 8 locations; sites YK, TL, AA, PK, MN, LN, XN and UY, see Extended Data Fig. 1).
Extended Data Figure 3 Patterns of genetic differentiation among eight sample sites where at least four individuals were genotyped.
The thickness of the lines connecting locations is inversely proportional to the FST values between two given locations. Purple lines connect each population with its closest neighbour around the ring. FST pairwise values ranged from 0.034 to 0.303.
Extended Data Figure 4 Plots of genetic admixture proportions (Q) according to additional values of K.
Values of Q were inferred by the software STRUCTURE30. Colours are in agreement with those used to label each subspecies at the bottom of this figure.
Supplementary information
Supplementary Information File 1
Genotypes of 95 greenish warblers for a selected set of 2,334 SNPs in GenAlex format. Individual IDs are displayed in the first column and SNP marker genotypes are displayed in two columns. Numerical codes and colours represent the four different nucleotide bases (A=101=red; C=102=blue; G=103=yellow; T=104=green). The bottom row indicates the chromosome (using the zebra finch genome assembly28 as reference) where each SNP marker is located. (XLSX 1543 kb)
Supplementary Information File 2
Genotypes of 4,018 polymorphic SNP markers across the greenish warbler range in a set of individuals sampled from Kyrgyzstan (site AA) to Nepal (site LN) that encompasses the entire sampled range of P.t ludlowi. The file is in GenAlex format, as described in SI File 1. (XLSX 1749 kb)
Supplementary Information File 3
Genotypes of greenish warblers for a set of highly differentiated markers (FST >0.85), grouped by chromosome, between viridanus and plumbeitarsus in GenAlex format. P. t. plumbeitarsus individuals with large chunks of DNA introgressing from viridanus are highlighted in yellow. Principal Coordinate Analyses for individual chromosomes are shown in the worksheet labeled as "PCAs". Here, individuals showing mixed ancestry are emphasized with blue squares. The ID of each individual is also given besides each PCA plot. Chromosomes without significant signal of introgression are also included for comparative purposes (e.g. ChrZ, Chr28 and Chr15). The position of each marker, using the zebra finch genome assembly as reference28, is indicated in the worksheet labeled as "Location". (XLSX 948 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Alcaide, M., Scordato, E., Price, T. et al. Genomic divergence in a ring species complex. Nature 511, 83–85 (2014). https://doi.org/10.1038/nature13285
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13285
This article is cited by
High heterogeneity in genomic differentiation between phenotypically divergent songbirds: a test of mitonuclear co-introgression
Heredity (2023)
Development and testing of diagnostic molecular markers to differentiate among three species of Salvelinus: Arctic char, Dolly Varden, and Bull trout
Conservation Genetics Resources (2023)
Distant but related: genetic structure in the circum-boreal bumblebee Bombus jonellus (Kirby, 1802)
Polar Biology (2021)
Climate Change Impacts on Himalayan Biodiversity: Evidence-Based Perception and Current Approaches to Evaluate Threats Under Climate Change
Journal of the Indian Institute of Science (2021)
Species limits and biogeography of Rhynchospiza sparrows
Journal of Ornithology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.