Abstract
We respond to a comment by Allmon WD (2016), who attempted to demonstrate that species are biologically ‘real’ as justification for retaining the terms ‘anagenesis’ and ‘cladogenesis’, which we argue are not necessary for the study of evolutionary biology (Vaux F, Trewick SA & Morgan-Richards M, 2016). Here, we summarize a wealth of literature demonstrating that supposedly separate species introgress frequently, and we clarify that evolutionary lineage-splitting with genotypic and phenotypic divergence (speciation) is not the same as taxonomic classification. The usefulness of the terms anagenesis and cladogenesis requires agreement on their meaning, and this debate reflects a wider dilemma in academic communication: whether to use imprecisely defined jargon or longer sentences with simple words. We favour the latter, because biological evolution operates under straightforward and generalizable principles that should not require complicated descriptions, especially when its study requires collaboration among many disciplines.
Introduction
In a previous review appearing in the Biological Journal of the Linnean Society (Vaux, Trewick & Morgan-Richards, 2016), we considered the usage and meaning of the terms ‘anagenesis’ and ‘cladogenesis’. We observed that the meaning of these terms has changed over time, and that modern usage is highly varied across disciplines and also often ambiguous. We concluded that the terms anagenesis and cladogenesis were not needed to describe evolution or species classification, and that they potentially hamper communication between disciplines. For example, some studies define ‘anagenesis’ as evolutionary change within a species (Johnson et al., 2012; Hunt, 2013; Lister, 2013), whereas others consider the term to be synonymous with gradualism (Ricklefs, 2004; Theriot et al., 2006; Mattila & Bokma, 2008; Pearson & Ezard, 2014). Variation in usage between disciplines is obvious. For example, many palaeontologists only recognize ‘anagenesis’ when morphospecies do not coexist temporally (Gould, 2001; MacFadden et al., 2012); whereas it is common for biogeographers to consider contemporary but geographically isolated lineages as examples of anagenetic speciation (Rosindell & Phillimore, 2011; Patiño et al., 2014; Valente, Etienne & Phillimore, 2014).
The mode of evolution may be anagenetic if the [first appearance] of the descendant coincides with the [last appearance] of the ancestor within the bounds of the dating precision. (Strotz & Allen, 2013)
A common mode of speciation in ocean islands is by anagenesis, wherein an immigrant arrives and, through time, transforms by mutation, recombination, and drift into a morphologically and genetically distinct species. (López-Sepúlveda et al., 2015)
In a response, Allmon (2016) agrees with much of our review but promotes the treatment of species as being biologically real (Allmon, 2016). This contrasts with our approach of treating species classification as arbitrary segments of an evolutionary lineage (Vaux et al., 2016). We welcome the recognition (Allmon, 2016) that ‘change’ and ‘branching’ are suitable substitutes for anagenesis and cladogenesis in many discussions of evolution (Simpson, 1944; Simpson 1953).
Species and Genetic Introgression
It is not necessary for us to reiterate the thorough exploration of the nature of species and their delimitation (Darwin, 1859; Mayr, 1942; Ghiselin, 1974; Burger, 1975; Mahner, 1993; Mallet, 1995; de Queiroz, 1998; Sites & Marshall, 2003; Hey, 2006; Konstantinidis et al., 2006; Dubois, 2011) because it does not actually address our criticism of ‘anagenesis’ and ‘cladogenesis’, nor does it demonstrate the necessity of the terms. Nonetheless, we do favour the acceptance that species are essentially arbitrary constructs because no concept can be universally and consistently applied to evolving biota (Vaux et al., 2016). In doing so, we follow the simple and well accepted fact identified by Darwin (1859) that species cannot be immutable at the same time as also evolving. Specifically, we observe that, although some species appear to coincide with in vogue concepts, every species is an arbitrary segment of an evolutionary lineage in time (de Queiroz, 1998, 2007; Vaux et al., 2016). We agree with Allmon (2016) that species can be established on the biologically real phenomena of evolutionary lineages [a line of descent of evolutionary units (organisms, replicators)], although the delimitation of a segment (especially in time) remains arbitrary (de Queiroz, 2011). This is because divergence and lineage-splitting are not always concordant and partitions of variation among evolutionary lineages are ultimately of subjective interest to biologists. Practically, one can rarely identify a discrete origin of a species (if such an event ever occurs) and, theoretically, speciation is an infinite process referring to change among related evolutionary lineages.
We agree with a source cited by Allmon (2016) that ‘a generally applicable concept of a species does not yet exist’ (Marie Curie Speciation Network, 2012). The claim that there is a consensus for the definition of a species for ‘at least the biparental animal part of [the living world]’ (Allmon, 2016) is readily falsified (see below) and the need for such a qualifier exposes the inadequacy of the assertion. A unifying concept cannot apply to only a subset of lineages in evolutionary time. Allmon (2016) promotes the view that species are biologically real and, although some taxonomic species are closer representations of evolutionary lineages than others (Rieseberg, Wood & Baack, 2006), problematic organisms remain abundant (Burger, 1975; Diamond, 1992; Berger & Ogielska, 1994; Domingo et al., 1995; Konstantinidis et al., 2006; Rieseberg et al., 2006; Chan et al., 2012; Fuchs et al., 2015).
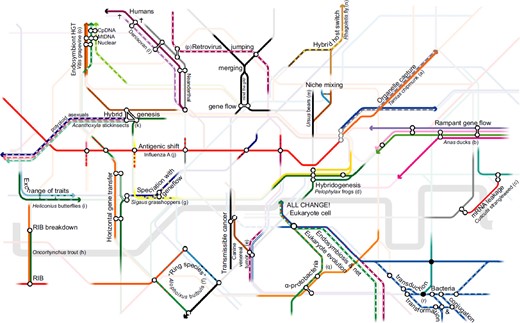
Despite previous reviews (Anderson & Stebbins, 1954; Mallet, 2007; Harrison, 2012; Abbott et al., 2013), it appears that the impact of introgression upon speciation and taxonomic classification is not fully appreciated. Introgression originates from two sources: reproduction (or vertical gene transfer) and horizontal gene transfer (Fig. 1). Although hybridization involving reproduction between members of separate lineages sometimes results in nonviable or infertile offspring (Wishart et al., 1988; Allen & Short, 1997; Rieseberg, 1997, Davis et al., 2015), this is not always the case (Burger, 1975; Rieseberg, 1997; Manos, Doyle & Nixon, 1999; Petit et al., 2003; Morgan-Richards et al., 2004; Trewick, Morgan-Richards & Chapman, 2004), even among biparental sexual animals (Derr et al., 1991; Rhymer, Williams & Braun, 1994; Schwarz et al., 2005; Gelberg, 2009; Kraus et al., 2012; The Heliconius Genome Consortium, 2012; Cahill et al., 2013; Bull & Sunnucks, 2014; Dowle, Morgan-Richards & Trewick, 2014; Liu et al., 2014; Prüfer et al., 2014; Fuchs et al., 2015; Good et al., 2015; Mckean, Trewick & Morgan-Richards, 2016; Morgan-Richards et al., 2016; Fig 1). Even notoriously infertile first-generation hybrids such as mules (Equus) can occasionally be fertile (Allen & Short, 1997), as can lineages that require sexual stimuli or gametes of another lineage (Berger & Ogielska, 1994; Ragghianti et al., 2007), and hybridization among distantly related organisms is well documented (Rieseberg & Willis, 2007; Rothfels et al., 2015). Furthermore, hybrid reproduction can be a source of hybrid vigour and it can transfer highly advantageous traits (The Heliconius Genome Consortium, 2012).
Figure 1.
Hybrid reproduction (vertical gene transfer) and horizontal gene transfer (HGT) result in frequent introgression among evolutionary lineages, which reveals that putative taxonomic species do not remain separate and that their delimitation is ultimately subjective. Here, we illustrate some of the range of processes that have so far been identified, citing examples in the primary and review literature (letter codes a - r refer to references listed in Supporting Information Table S1, and many other examples are provided therein). Re-purposing an unrelated network demonstrates that modes of introgression are so prolific and diverse that almost any example can be illustrated by an arbitrary selection of intersecting lines. Note that the network relationships demonstrate gene flow but not phylogeny or a scaled representation of change through time.
Horizontal gene transfer (HGT) has had a significant impact over evolutionary time in all major clades of life. Models for the evolution of the eukaryotic cell rely upon HGT and subsequent genetic introgression (Margulis, Dolan & Guerrero, 2000; Georgiades & Raoult, 2011, 2012), and abundant evidence demonstrates that organellar DNA is continuously transferred to the nucleus (Blanchard & Lynch, 2000; Stegemann et al., 2003), as well as between organelles (Goremykin et al., 2009). Other prokaryotic endosymbionts (organisms within the cells of another) are also absorbed (Gonella et al., 2015) and undergo HGT (Kondo et al., 2002; Husnik et al., 2013; Sloan et al., 2014; Wybouw et al., 2014), and viruses facilitate HGT between themselves and eukaryotic host genomes (Bejarano et al., 1996; Löwer, Löwer & Kurth, 1996; Mallet et al., 2004; Carrat & Flahault, 2007; Herniou et al., 2013; Gasmi et al., 2015). Even the most reproductively discrete, biparental, sexual animals are therefore continuously introgressing with DNA of prokaryotic and viral origin. HGT is near-constant in bacteria via direct cell-to-cell exchange, indirect environmental exchange between cells, and indirect exchange between cells via viral infection (Ochman, Lawrence & Groisman, 2000; Krebes et al., 2014). In many mutualistic and parasitic situations, nonvectored HGT involves all combinations of animals, bacteria, fungi, and plants, including both nuclear and organellar DNA (Vaughn et al., 1995; Groth, Hansen & Piškur, 1999; Davis & Wurdack, 2004; Woloszynska et al., 2004; Hall, Brachat & Dietrich, 2005; Moran & Jarvik, 2010; Yoshida et al., 2010; Acuña et al., 2012; Kim et al., 2014; Nikolaidis, Doran & Cosgrove, 2014; Wybouw et al., 2014). HGT is observed between animal hosts and transmissible cancers (Metzger et al., 2016; Strakova et al., 2016) and syncytial growth (nuclei sharing among cells) in fungi also provides the potential for HGT and viable interspecies genetic mosaics (in sensuRoper et al., 2013). These genetic exchanges often produce functional genes (Mallet et al., 2004; Nikolaidis et al., 2014), and associated traits often have the potential to be significantly advantageous and are of clear taxonomic interest (Bock, 2010; Moran & Jarvik, 2010; Herniou et al., 2013; Nikolaidis et al., 2014; Crisp et al., 2015; Gasmi et al., 2015). Resulting changes in the evolutionary trajectory of a lineage affect the overall pattern of lineage-splitting and divergence among populations, meaning that introgression does not merely result in gene tree heterogeneity. A plethora of examples illustrate how reproduction and HGT maintain introgression and unclear boundaries for species classification (Fig. 1); species do not ‘maintain their separateness’ (Allmon, 2016).
In some ways, we and Allmon are speaking past one another because perceptions of the status of species are sensitive to the resolution at which they are observed. At the scale typically used to investigate trends in biodiversity, species can appear coherent and separate. Most taxonomic work depends on arbitrary distinctions made by experts with the primary objective of defining distinct units. However, at a closer range where lineage-splitting and divergence are studied in detail, it often becomes apparent that such coherence is superficial. Under most definitions (Aze et al., 2013; Lister, 2013; Strotz & Allen, 2013), it is this scale of lineage-splitting at which periods of anagenesis and cladogenesis are defined, and thus where problems arise. Similar scale differences also affect the study of topics such as evolutionary stasis, where a trait can appear morphologically static over long periods of time but less so over a shorter time period with more frequent sampling intervals (Hunt, 2012).
‘When I use a word’, Humpty Dumpty said in a rather scornful tone, ‘it means just what I choose it to mean – neither more nor less’.
(Lewis Carroll, 1871 in Through The Looking Glass)
[also aptly quoted in Harrison, 2012]
Allmon (2016) conflates species classification (and delimitation) with speciation by suggesting that we are not interested in studying speciation. Although seemingly an arid enterprise, clarification of terms used in evolutionary biology is needed for the intelligent exploration of biology. We explicitly stated that the classification (and observation) of a species depends upon divergence-based factors and the hypothesis of interest (Vaux et al., 2016). What this means is that the origination of species as a classified taxon is arbitrary, although the process of lineage-splitting and divergence that creates the diversity used to describe it is biologically real (and interesting). When most evolutionary biologists refer to ‘speciation’, we assume that they mean the latter process, and not the pedantic and arbitrary delimitation of a taxon. The process is of interest because it considers the biological evidence available (genetic variation, phenotypic variation, selection), whereas taxonomy is deciding when and how to assign names based, usually on a subset of that evidence. The fact that we treat a species as an arbitrary concept does not prevent hypothesis testing, the study of lineage-splitting, divergence or diversification rates, or investigation of the fossil record (Darwin, 1859).
Anagenesis and Cladogenesis
There are many instances where palaeontological evidence provides estimates of when lineage-splits must have occurred (e.g. Strotz & Allen, 2013; Pearson & Ezard, 2014; Kimura, Flynn & Jacobs, 2016) and we also agree that palaeontologically recognized species can be comparable to living taxa (even if this is difficult to demonstrate) on a lineage divided into segments in time (de Queiroz, 1998; Kimura et al., 2016). However, morphological crypsis leading to underestimation of diversity is not the only problem for the morphological identification of extinct species. The treatment of ‘estimates of species and speciation rates [as] minimum estimates’ (Allmon, 2016) is flawed because there are also cases of taxonomic over-splitting in palaeontology that leads to overestimation of diversity (Hills et al., 2012; Aze et al., 2013).
Despite lengthy discussion of species classification in the fossil record, Allmon (2016) does not define the terms or address the actual concern of our review: the ability to consistently define (and delineate in time) anagenesis (phyletic change) and cladogenesis (divergence concurrent with lineage-splitting) based on morphological evidence alone. Morphological divergence and lineage-splitting are not necessarily concordant. Even in palaeontological studies incorporating genetic data, estimates that utilize independent loci within a lineage will provide a range of dates (rather than a single estimate) for a lineage-split. This is problematic for the delimitation of anagenesis and cladogenesis because most palaeontological definitions assume their mutual exclusivity (Aze et al., 2013; Lister, 2013).
Never use a long word when a diminutive one will do.
(William Safire, 1979 in On Language, New York Times Magazine)
The claim that we have only demonstrated ‘disparate usage by a few modern authors’ (Allmon, 2016) is inaccurate because our review cited many recent papers that vary in the meaning given to anagenesis and cladogenesis (Mattila & Bokma, 2008; Drew & Barber, 2009; Catley, Novick & Shade, 2010; Dubois, 2011; Johnson et al., 2012; Pachut & Anstey, 2012; Aze et al., 2013; Bapst, 2013; Futuyma, 2013; Hunt, 2013; Podani, 2013; Strotz & Allen, 2013; Dynesius & Jansson, 2014; Pearson & Ezard, 2014; Patiño et al., 2014; Valente et al., 2014). For this contemporary variation to exist, the terms cannot have remained consistent ‘for more than half a century’, as Allmon (2016) suggests. We do not think this variation should be ignored because previous studies also discuss the problematic meaning of the terms (Benton & Pearson, 2001; Dubois, 2011) and also because textbooks and educational research demonstrate that definitions vary (Catley et al., 2010; Johnson et al., 2012; Futuyma, 2013), indicating that this ambiguity may be inherited by future scientists. If we follow the definition used by Simpson (1944), as suggested by Allmon (2016), why do we need multiple words for ‘branching’ and ‘phyletic change’? What is the necessity of redundant terminology (likewise with ‘tokogenesis’ for gene flow; Allmon, 2016)?
Conclusions
Ultimately, the necessity of terms such as anagenesis and cladogenesis reflects a wider problem in academic communication. Researchers will decide whether to use complex terminology (giving each term the meaning they choose) or longer sentences with simple words. Biological evolution fundamentally operates under the basic principles of variation, selection, and heritability, which can be effectively modelled using even simple descriptions such as the univariate breeder's equation (R = Sh2). Although this process generates rich complexity in nature, we consider that descriptions of biological evolution need not require complex and alienating language. We do not expect everyone to agree with our views on anagenesis and cladogenesis, although we hope it can at least be agreed that the terms in their current state are problematic for the communication of science and, in future, authors should clearly express their definition of the terms or otherwise avoid them.
Acknowledgements
We are grateful for the constructive criticism and feedback from the reviewers Warren D. Allmon and William Miller III. We thank the Biological Journal of the Linnean Society and the editor John A. Allen for the opportunity to publish our response. This work was supported by the Royal Society of New Zealand Te Apārangi Marsden Fund grant (12-MAU-008).
References
Abbott
R
Albach
D
Ansell
S
Arntzen
JW
Baird
SJE
Bierne
N
Boughman
J
Brelsford
A
Buerkle
CA
Buggs
R
Butlin
RK
Dieckmann
U
Eroukhmanoff
F
Grill
A
Cahan
SH
Hermansen
JS
Hewitt
G
Hudson
AG
Jiggins
C
Jones
J
Keller
B
Marczewski
T
Mallet
J
Martinez-Rodriguez
P
Mӧst
M
Mullen
S
Nichols
R
Nolte
AW
Parisod
C
Pfennig
K
Rice
AM
Ritchie
MG
Seifert
B
Smadja
CM
Stelkens
R
Szymura
JM
Väinölä
R
Wolf
JBW
Zinner
D
.
2013
.
Hybridization and speciation
.
Journal of Evolutionary Biology
26
:
229
–
246
.
Acuña
R
Padilla
BE
Flórez-Ramos
C
Rubio
JD
Herrera
JC
Benavides
P
Lee
S
Yeats
TH
Egan
AN
Doyle
JJ
Rose
JKC
.
2012
.
Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee
.
Proceedings of the National Academy of Sciences of the United States of America
109
:
4197
–
4202
.
Allen
WR
Short
RV
.
1997
.
Interspecific and extraspecific pregnancies in Equids: anything goes
.
Journal of Heredity
88
:
384
–
392
.
Allmon
WD
.
2016
.
Species, lineages, splitting, and divergence: why we still need ‘anagenesis’ and ‘cladogenesis’
.
Biological Journal of the Linnean Society
doi: 10.1111/bij.12885
Anderson
E
Stebbins
GL
Jr .
1954
.
Hybridization as an evolutionary stimulus
.
Evolution
8
:
378
–
388
.
Aze
T
Ezard
THG
Purvis
A
Coxall
HK
Stewart
DRM
Wade
BS
Pearson
PN
.
2013
.
Identifying anagenesis and cladogenesis in the fossil record
.
Proceedings of the National Academy of Sciences of the United States of America
110
:
E2946
.
Bapst
DW
.
2013
.
When can clades be potentially resolved with morphology?
PLoS ONE
8
:
e62312
.
Bejarano
ER
Khashoggi
A
Witty
M
Lichtenstein
C
.
1996
.
Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution
.
Proceedings of the National Academy of Sciences of the United States of America
93
:
756
–
764
.
Benton
MJ
Pearson
PN
.
2001
.
Speciation in the fossil record
.
Trends in Ecology and Evolution
16
:
405
–
411
.
Berger
L
Ogielska
M
.
1994
.
Spontaneous haploid–triploid mosaicism in the progeny of a Rana kl. esculenta female and Rana lessonae males
.
Amphibia-Reptilia
15
:
143
–
152
.
Blanchard
JL
Lynch
M
.
2000
.
Organellar genes: why do they end up in the nucleus?
Trends in Genetics
16
:
315
–
320
.
Bock
R
.
2010
.
The give-and-take of DNA: horizontal gene transfer in plants
.
Trends in Plant Science
15
:
11
–
22
.
Bull
JK
Sunnucks
P
.
2014
.
Strong genetic structuring without assortative mating or reduced hybrid survival in an onychophoran in the Tallaganda State Forest region, Australia
.
Biological Journal of the Linnean Society
111
:
589
–
602
.
Burger
WC
.
1975
.
The species concept in Quercus
.
Taxon
24
:
45
–
50
.
Cahill
JA
Green
RE
Fulton
TL
Stiller
M
Jay
F
Ovsyanikov
N
Salamzade
R
St. John
J
Stirling
I
Slatkin
M
Shapiro
B
.
2013
.
Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution
.
PLOS Genetics
9
:
e1003345
.
Carrat
F
Flahault
A
.
2007
.
Influenza vaccine: the challenge of antigenic shift
.
Vaccine
25
:
6852
–
6862
.
Catley
KM
Novick
LR
Shade
CK
.
2010
.
Interpreting evolutionary diagrams: when topology and process conflict
.
Journal of Research in Science Teaching
47
:
861
–
882
.
Chan
JZ-M
Halachev
MR
Loman
NJ
Constantinidou
C
Pallen
MJ
.
2012
.
Defining bacterial species in the genomic era: insights from the genus Acinetobacter
.
BMC Microbiology
12
:
302
.
Crisp
A
Boschetti
C
Perry
M
Tunnacliffe
A
Micklem
G
.
2015
.
Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes
.
Genome Biology
16
:
50
.
Darwin
CR
.
1859
.
On the origin of species
.
London
:
John Murray
.
Davis
CC
Wurdack
KJ
.
2004
.
Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales
.
Science
305
:
676
–
678
.
Davis
BW
Seabury
CM
Brashear
WA
Li
G
Roelke-Parker
M
Murphy
WJ
.
2015
.
Mechanisms underlying mammalian hybrid sterility in two feline interspecies models
.
Molecular Biology and Evolution
32
:
2534
–
2546
.
Derr
JN
Hale
DW
Ellsworth
DL
Bickham
JW
.
1991
.
Fertility in an F1 hybrid of white-tailed deer (Odocoileus virginianus) × mule deer (O. hemionus)
.
Journal of Reproduction and Fertility
93
:
111
–
117
.
Diamond
JM
.
1992
.
Horrible plant species
.
Nature
360
:
627
–
628
.
Domingo
E
Holland
JJ
Biebricher
C
Eigen
M
.
1995
. Quasi-species: the concept and the word. In:
Gibbs
AJ
Calisher
CH
García-Arenal
F
, eds.
Molecular basis of virus evolution
.
Cambridge
:
Cambridge University Press
,
181
–
191
.
Dowle
EJ
Morgan-Richards
M
Trewick
SA
.
2014
.
Morphological differentiation despite gene flow in an endangered grasshopper
.
BMC Evolutionary Biology
14
:
216
.
Drew
J
Barber
PH
.
2009
.
Sequential cladogenesis of the reef fish Pomacentrus moluccensis (Pomacentridae) supports the peripheral origin of marine biodiversity in the Indo-Australian archipelago
.
Molecular Phylogenetics and Evolution
53
:
335
–
339
.
Dubois
A
.
2011
.
Species and ‘strange species’ in zoology: do we need a ‘unified concept of species’?
Comptes Rendus Palevol
10
:
77
–
94
.
Dynesius
M
Jansson
R
.
2014
.
Persistence of within-species lineages: a neglected control of speciation rates
.
Evolution
68
:
923
–
934
.
Fuchs
J
Ericson
PGP
Bonillo
C
Couloux
A
Pasquet
E
.
2015
.
The complex phylogeography of the Indo-Malayan Alophoixus bulbuls with the description of a putative new ring species complex
.
Molecular Ecology
24
:
5460
–
5474
.
Futuyma
DJ
.
2013
.
Evolution, 3rd edn
.
Sunderland, MA
:
Sinaeur Associates
.
Gasmi
L
Boulain
H
Gauthier
J
Hua-Van
A
Musset
K
Jakubowska
AK
Aury
J-M
Volkoff
A-N
Huguet
E
Herrero
S
Drezen
J-M
.
2015
.
Recurrent domestication by Lepidoptera of genes from their parasites mediated by Bracoviruses
.
PLOS Genetics
11
:
e1005470
.
Gelberg
HB
.
2009
.
Purkinje fiber dysplasia (histiocytoid cardiomyopathy) with ventricular noncompaction in a savannah kitten
.
Veterinary Pathology
46
:
693
–
697
.
Georgiades
K
Raoult
D
.
2011
.
The rhizome of Reclinomonas americana, Homo sapiens, Pediculus humanus and Saccharomyces cerevisiae mitochondria
.
Biology Direct
6
:
55
.
Georgiades
K
Raoult
D
.
2012
.
How microbiology helps define the rhizome of life
.
Frontiers in Cellular and Infection Microbiology
2
:
60
.
Ghiselin
MT
.
1974
.
A radical solution to the species problem
.
Systematic Zoology
23
:
536
–
544
.
Gonella
E
Pajoro
M
Marzorati
M
Crotti
E
Mandrioli
M
Pontini
M
Bulgari
D
Negri
I
Sacchi
L
Chouaia
B
Daffonchio
D
Alma
A
.
2015
.
Plant-mediated interspecific horizontal transmission of an intracellular symbiont in insects
.
Scientific Reports
5
:
15811
.
Good
JM
Vanderpool
D
Keeble
S
Bi
K
.
2015
.
Negligible nuclear introgression despite complete mitochondrial capture between two species of chipmunks
.
Evolution
69
:
1961
–
1972
.
Goremykin
VV
Salamini
F
Valesco
R
Viola
R
.
2009
.
Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer
.
Molecular Biology and Evolution
26
:
99
–
110
.
Gould
SJ
.
2001
. The interrelationship of speciation and punctuated equilibrium. In:
Cheetham
AH
Jackon
JBC
Lidgard
S
McKinney
FK
, eds.
Evolutionary patterns: growth, form, and tempo in the fossil record
.
Chicago, IL
:
University of Chicago Press
,
207
–
208
.
Groth
C
Hansen
J
Piškur
J
.
1999
.
A natural chimeric yeast containing genetic material from three species
.
International Journal of Systematic Bacteriology
49
:
1933
–
1938
.
Hall
C
Brachat
S
Dietrich
FS
.
2005
.
Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae
.
Eukaryotic Cell
4
:
1102
–
1115
.
Harrison
RG
.
2012
.
The language of speciation
.
Evolution
66
:
3643
–
3657
.
Herniou
EA
Huguet
E
Thézé
J
Bézier
A
Periquet
G
Drezen
J-M
.
2013
.
When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses
.
Philosophical Transactions of the Royal Society Series B, Biological Sciences
368
:
20130051
.
Hey
J
.
2006
.
On the failure of modern species concepts
.
Trends in Ecology and Evolution
21
:
447
–
450
.
Hills
SFK
Crampton
JS
Trewick
SA
Morgan-Richards
M
.
2012
.
DNA and morphology unite two species and 10 million year old fossils
.
PLoS ONE
12
:
e52083
.
Hunt
G
.
2012
.
Measuring rates of phenotypic evolution and inseparability of tempo and mode
.
Paleobiology
38
:
351
–
373
.
Hunt
G
.
2013
.
Testing the link between phenotypic evolution and speciation: an integrated palaeontological and phylogenetic analysis
.
Methods in Ecology and Evolution
4
:
714
–
723
.
Husnik
F
Nikoh
N
Koga
R
Ross
L
Duncan
RP
Fujie
M
Tanaka
M
Satoh
N
Bachtrog
D
Wilson
ACC
von Dohlen
CD
Fukatsu
T
McCutcheon
JP
.
2013
.
Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis
.
Cell
153
:
1567
–
1578
.
Johnson
NA
Smith
JJ
Pobiner
B
Schrein
C
.
2012
.
Why are chimps still chimps?
The American Biology Teacher
74
:
74
–
80
.
Kim
G
LeBlanc
ML
Wafula
EK
dePamphilis
CW
Westwood
JH
.
2014
.
Genomic-scale exchange of mRNA between a parasitic plant and its hosts
.
Science
345
:
808
–
811
.
Kimura
Y
Flynn
LJ
Jacobs
LL
.
2016
.
A palaeontological case study for species delimitation in diverging fossil lineages
.
Historical Biology
28
:
189
–
198
.
Kondo
N
Nikoh
N
Ijichi
N
Shimada
M
Fukatsu
T
.
2002
.
Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect
.
Proceedings of the National Academy of Sciences of the United States of America
99
:
14281
–
14285
.
Konstantinidis
KT
Ramette
A
Tiedje
JM
.
2006
.
The bacterial species definition in the genomic era
.
Philosophical Transactions of the Royal Society Series B, Biological Sciences
361
:
1929
–
1940
.
Kraus
RHS
Kerstens
HHD
van Hooft
P
Mergens
H
Elmberg
J
Tsvey
A
Sartakov
D
Soloviev
SA
Crooijmans
RPMA
Groenen
MAM
Ydenberg
RC
Prins
HHT
.
2012
.
Widespread horizontal genomic exchange does not erode species barriers among sympatric ducks
.
BMC Evolutionary Biology
12
:
45
.
Krebes
J
Didelot
X
Kennemann
L
Suerbaum
S
.
2014
.
Bidirectional genomic exchange between Helicobacter pylori strains from a family in Coventry, United Kingdom
.
International Journal of Medical Microbiology
304
:
1135
–
1146
.
Lister
AM
.
2013
. Speciation and evolutionary trends in quaternary vertebrates. In:
Elias
S
Mock
C
, eds.
Encyclopedia of quaternary science, 2nd edn
.
Amsterdam
:
Elsevier
,
723
–
732
.
Liu
S
Lorenzen
ED
Fumagalli
M
Li
B
Harris
K
Xiong
Z
Zhou
L
Sand Koreneliussen
T
Somel
M
Babbitt
C
Wray
G
Li
J
He
W
Wang
Z
Fu
W
Xiang
X
Morgan
CC
Doherty
A
O'Connell
MJ
Zhang
G
Nielsen
R
Willerslev
E
Wang
J
.
2014
.
Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears
.
Cell
157
:
785
–
794
.
López-Sepúlveda
P
Takayama
K
Greimler
J
Crawford
DJ
Peñailillo
P
Baeza
M
Ruiz
E
Kohl
G
Tremetsberger
K
Gatica
A
Letelier
L
Novoa
P
Novak
J
Stuessy
TF
.
2015
.
Progressive migration and anagenesis in Drimys conferifolia of the Juan Fernández Archipelago, Chile
.
Journal of Plant Research
128
:
73
–
90
.
Löwer
R
Löwer
J
Kurth
R
.
1996
.
The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences
.
Proceedings of the National Academy of Sciences of the United States of America
93
:
5177
–
5184
.
MacFadden
BJ
Oviedo
LH
Seymour
GM
Ellis
S
.
2012
.
Fossil horses, orthogenesis, and communicating evolution in museums
.
Evolution: Education and Outreach
5
:
29
–
37
.
Mahner
M
.
1993
.
What is a species? A contribution to the never ending species debate in biology
.
Journal of General Philosophy of Science
24
:
103
–
126
.
Mallet
J
.
1995
.
A species definition for the modern synthesis
.
Trends in Ecology and Evolution
10
:
294
–
299
.
Mallet
J
.
2007
.
Hybrid speciation
.
Nature
446
:
279
–
283
.
Mallet
F
Bouton
O
Prudhomme
S
Cheynet
V
Oriol
G
Bonnaud
B
Lucotte
G
Duret
L
Mandrand
B
.
2004
.
The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology
.
Proceedings of the National Academy of Sciences of the United States of America
101
:
1731
–
1736
.
Manos
PS
Doyle
JJ
Nixon
KC
.
1999
.
Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae)
.
Molecular Phylogenetics and Evolution
12
:
333
–
349
.
Margulis
L
Dolan
MF
Guerrero
R
.
2000
.
The chimeric eukaryote: origin of the nucleus from the karyomastigont in amitochondriate protists
.
Proceedings of the National Academy of Sciences of the United States of America
97
:
6954
–
6959
.
Mattila
TM
Bokma
F
.
2008
.
Extant mammal body masses suggest punctuated equilibrium
.
Philosophical Transactions of the Royal Society Series B, Biological Sciences
275
:
2195
–
2199
.
Mayr
E
.
1942
.
Systematics and the origin of species from the viewpoint of a zoologist
.
New York, NY
:
Columbia University Press
.
Mckean
NE
Trewick
SA
Morgan-Richards
M
.
2016
.
Little or no gene flow despite F1 hybrids at two interspecific contact zones
.
Ecology and Evolution
6
:
2390
–
2404
.
Moran
NA
Jarvik
T
.
2010
.
Lateral transfer of genes from fungi underlies carotenoid production in aphids
.
Science
328
:
624
–
627
.
Metzger
MJ
Villalba
A
Carballal
MJ
Iglesias
D
Sherry
J
Reinisch
C
Muttray
AF
Baldwin
SA
Goff
SP
.
2016
.
Widespread transmission of independent cancer lineages within multiple bivalve species
.
Nature
534
:
705
.
Morgan-Richards
M
Trewick
SA
Chapman
HM
Krahulcova
A
.
2004
.
Interspecific hybridization among Hieracium species in New Zealand: evidence from flow cytometry
.
Heredity
93
:
34
–
42
.
Morgan-Richards
M
Hills
SKF
Biggs
PJ
Trewick
SA
.
2016
.
Sticky genomes: using NGS to test hybrid speciation hypothesis
.
PLoS ONE
11
:
e0154911
.
Network MCS
.
2012
.
What do we need to know about speciation?
Trends in Ecology and Evolution
27
:
27
–
39
.
Nikolaidis
N
Doran
N
Cosgrove
DJ
.
2014
.
Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion
.
Molecular Biology and Evolution
31
:
376
–
386
.
Ochman
H
Lawrence
JG
Groisman
EA
.
2000
.
Lateral gene transfer and the nature of bacterial innovation
.
Nature
405
:
299
–
304
.
Pachut
JF
Anstey
RL
.
2012
.
Rates of anagenetic evolution and selection intensity in Middle and Upper Ordovician species of the bryozoan genus Peronopora
.
Paleobiology
38
:
403
–
423
.
Patiño
J
Carine
M
Fernández-Palacios
JM
Otto
R
Schaefer
H
Vanderpoorten
A
.
2014
.
The anagenetic world of spore-producing land plants
.
New Phytologist
201
:
305
–
311
.
Pearson
PN
Ezard
THG
.
2014
.
Evolution and speciation in the Eocene planktonic foraminifer Turborotalia
.
Paleobiology
40
:
130
–
143
.
Petit
RJ
Bodénès
C
Ducousso
A
Roussel
G
Kremer
A
.
2003
.
Hybridization as a mechanism of invasion in oaks
.
New Phytolologist
161
:
151
–
164
.
Podani
J
.
2013
.
Tree thinking, time and topology: comments on the interpretation of tree diagrams in evolutionary/phylogenetic systematics
.
Cladistics
29
:
315
–
327
.
Prüfer
K
Racimo
F
Patterson
N
Jay
F
Sankararaman
S
Sawyer
S
Heinze
A
Renaud
G
Sudmant
PH
de Filippo
C
Li
H
Mallick
S
Dannemann
M
Fu
Q
Kircher
M
Kuhlwilm
M
Lachmann
M
Meyer
M
Ongyerth
M
Siebauer
M
Theunert
C
Moorjani
P
Pickrell
J
Mullikin
JC
Vohr
SH
Green
RE
Hellmann
I
Johnson
PLF
Blanche
H
Cann
H
Kitzman
JO
Shendure
J
Eichler
EE
Lein
ES
Bakken
TE
Golovanova
LV
Doronichev
VB
Shunkov
MV
Derevianko
AP
Viola
B
Slatkin
M
Reich
D
Kelso
J
Pääbo
S
.
2014
.
The complete genome sequence of a Neanderthal from the Atlai Mountains
.
Nature
505
:
43
–
49
.
de Queiroz
K
.
1998
. The general lineage concept of species, species criteria, and the process of speciation. In:
Howard
DJ
Verlocher
SH
, eds.
Endless forms: species and speciation
.
Oxford
:
Oxford University Press
,
57
–
75
.
de Queiroz
K
.
2007
.
Species concepts and species delimitation
.
Systematic Biology
56
:
879
–
886
.
de Queiroz
K
.
2011
.
Branches in the lines of descent: Charles Darwin and the evolution of the species concept
.
Biological Journal of the Linnean Society
103
:
19
–
35
.
Ragghianti
M
Bucci
S
Marracii
S
Casola
C
Mancino
G
Hotz
H
Geux
G-D
Plötner
J
Uzzell
T
.
2007
.
Gametogenesis of intergroup hybrids of hemiclonal frogs
.
Genetical Research
89
:
39
–
45
.
Rhymer
JM
Williams
MJ
Braun
MJ
.
1994
.
Mitochondrial analysis of gene flow between New Zealand mallards (Anas platyrhynchos) and grey ducks (A. superciliosa)
.
The Auk
111
:
970
–
978
.
Ricklefs
RE
.
2004
.
Cladogenesis and morphological diversification in passerine birds
.
Nature
430
:
338
–
341
.
Rieseberg
LH
.
1997
.
Hybrid origins of plant species
.
Annual Review of Ecology and Systematics
28
:
359
–
389
.
Rieseberg
LH
Willis
JH
.
2007
.
Plant speciation
.
Science
317
:
910
–
914
.
Rieseberg
LH
Wood
TE
Baack
EJ
.
2006
.
The nature of plant species
.
Nature
440
:
524
–
527
.
Roper
M
Simonin
A
Hickey
PC
Leeder
A
Glass
NL
.
2013
.
Nuclear dynamics in a fungal chimera
.
Proceedings of the National Academy of Sciences of the United States of America
110
:
12875
–
12880
.
Rosindell
J
Phillimore
AB
.
2011
.
A unified model of island biogeography sheds light on the zone of radiation
.
Ecology Letters
14
:
552
–
560
.
Rothfels
CJ
Johnson
AK
Hovenkamp
PH
Swofford
DL
Roskam
HC
Fraser-Jenkins
CR
Windham
MD
Pryer
KM
.
2015
.
Natural hybridization between genera that diverged from each other approximately 60 million years ago
.
American Naturalist
185
:
433
–
442
.
Schwarz
D
Matta
BM
Shakir-Botteri
NL
McPheron
BA
.
2005
.
Host shift to an invasive plant triggers rapid animal hybrid speciation
.
Nature
436
:
546
–
549
.
Simpson
GG
.
1953
.
Major features of evolution
.
New York, NY
:
Columbia University Press
.
Simpson
GG
.
1944
.
Tempo and mode in evolution
.
New York, NY
:
Columbia University Press
.
Sites
JW
Marshall
JC
.
2003
.
Delimiting species: a Renaissance issue in systematic biology
.
Trends in Ecology and Evolution
18
:
462
–
470
.
Sloan
DB
Nakabachi
A
Richards
S
Qu
J
Canchi Murali
S
Gibbs
RA
Moran
NA
.
2014
.
Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects
.
Molecular Biology and Evolution
31
:
857
–
871
.
Stegemann
S
Hartmann
S
Ruf
S
Bock
R
.
2003
.
High-frequency gene transfer from the chloroplast genome to the nucleus
.
Proceedings of the National Academy of Sciences of the United States of America
100
:
8828
–
8833
.
Strakova
A
Leathlobhair
MN
Wang
G-D
Yin
T-T
Airikkala-Otter
I
Allen
JL
Allum
KM
Bansse-Issa
L
Bisson
JL
Domracheva
AC
de Castro
KF
Corrigan
AM
Cran
HR
Crawford
JT
Cutter
SM
Keenan
LD
Donelan
EM
Faramade
IA
Reynoso
EF
Fotopoulou
E
Fruean
SN
Gallardo-Arrieta
F
Glebova
O
Häfelin Manrique
R
Henriques
JJGP
Ignatenko
N
Koenig
D
Lanza-Perea
M
Lobetti
R
Lopez Quintana
AM
Losfelt
T
Marino
G
Martincorena
I
Martínez Castañeda
S
Martínez-López
MF
Meyer
M
Nakanwagi
B
De Nardi
AB
Neunzig
W
Nixon
SJ
Onsare
MM
Ortega-Pacheco
PM
Pye
RJ
Reece
JF
Rojas Gutierrez
J
Sadia
H
Schmeling
SK
Shamanova
O
Ssuna
RK
Steenland-Smit
AE
Svitich
A
Thoya Ngoka
I
Viţălaru
BA
de Vos
AP
de Vos
JP
Walkinton
O
Wedge
DC
Wehrle-Martinez
AS
van der Wel
MG
Widdowson
SAE
Murchison
EP
.
2016
.
Mitochondrial genetic diversity, selection and recombination in a canine transmissible cancer
.
Elife
5
:
e14552
.
Strotz
LC
Allen
PA
.
2013
.
Assessing the role of cladogenesis in macroevolution by integrating fossil and molecular evidence
.
Proceedings of the National Academy of Sciences of the United States of America
110
:
2904
–
2909
.
The Heliconius Genome Consortium
.
2012
.
Butterfly genome reveals promiscuous exchange of mimicry adaptations among species
.
Nature
487
:
94
–
98
.
Theriot
EC
Fritz
SC
Whitlock
C
Conley
DJ
.
2006
.
Late Quaternary rapid morphological evolution of an endemic diatom in Yellowstone Lake, Wyoming
.
Paleobiology
32
:
38
–
54
.
Trewick
SA
Morgan-Richards
M
Chapman
HM
.
2004
.
Chloroplast DNA diversity of Hieracium pilosella (Asteraceae) introduced to New Zealand: reticulation, hybridization, and invasion
.
American Journal of Botany
91
:
73
–
85
.
Valente
LM
Etienne
RS
Phillimore
AB
.
2014
.
The effects of island ontogeny on species diversity and phylogeny
.
Philosophical Transactions of the Royal Society B: Biological Sciences
281
:
20133227
.
Vaughn
JC
Mason
MT
Sper-Whitis
G
Kuhlman
P
Palmer
JD
.
1995
.
Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric Cox1 gene of Peperomia
.
Journal of Molecular Evolution
41
:
563
–
572
.
Vaux
F
Trewick
SA
Morgan-Richards
M
.
2016
.
Lineages, splits and divergence challenge whether the terms anagenesis and cladogenesis are necessary
.
Biological Journal of the Linnean Society
117
:
165
–
176
.
Wishart
WD
Hrudka
F
Schmutz
SM
Flood
PF
.
1988
.
Observations on spermatogenesis, sperm phenotype, and fertility in white-tailed x mule deer hybrids and a yak x cow hybrid
.
Canadian Journal of Zoology
66
:
1664
–
1671
.
Woloszynska
M
Bocer
T
Mackiewicz
P
Janska
H
.
2004
.
A fragment of chloroplast DNA was transferred horizontally, probably from non-eudicots, to mitochondrial genome of Phaseolus
.
Plant Molecular Biology
56
:
811
–
820
.
Wybouw
N
Dermauw
W
Tirry
L
Stevens
C
Grbić
M
Feyereisen
R
Van Leeuwen
T
.
2014
.
A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning
.
Elife
3
:
e02365
.
Yoshida
S
Maruyama
S
Nozaki
H
Shirasu
K
.
2010
.
Horizontal gene transfer by the parasitic plant Striga hermonthica
.
Science
328
:
1128
.
Supporting Information
Additional Supporting Information may be found online in the supporting information tab for this article:
Table S1. Primary and review references for evidence of particular modes of genetic introgression via reproduction [vertical gene transfer (VGT)] and horizontal gene transfer (HGT), most of which are illustrated by single examples in Fig. 1. Only a small amount of the available literature is listed, and we deliberately focus upon examples from biparental sexual animals.
Author notes
© 2016 The Linnean Society of London, Biological Journal of the Linnean Society
PDF