Influenza Virology: Current Topics
"I recommend it for all virologists and public health scientists" (ASM Microbe)
"one of the best books currently available on Influenza virus research" (Google Books)
Publisher: Caister Academic Press
Edited by: Yoshihiro Kawaoka
School of Veterinary Medicine, University of Wisconsin-Madison, Madison, WI 53706, USAPages: viii + 368 + 5 colour plates
Hardback: Publication date: March 2006
ISBN: 978-1-904455-06-6
Price: GB £219 or US $319
Buy bookEbook: Publication date: March 2006
ISBN: 978-1-913652-31-9
Price: US $319
Buy ebookDOI: https://doi.org/10.21775/9781908230584
Customers who viewed this book also viewed:
Three times in the last century, influenza viruses have undergone major genetic changes resulting in global pandemics that had devastating effects. The most infamous pandemic was the Spanish Flu which affected up to 25% of the world population and is thought to have killed at least 40 million people in 1918-1919. More recently, two other influenza pandemics, the Asian Flu in 1957 and the Hong Kong Flu in 1968, killed millions of people worldwide. These caused severe disease, not only in the young and the elderly, who are usually very susceptible to influenza, but also among healthy younger persons. In 1997 and 2003 a new influenza A virus of H5N1 subtype emerged in Asia and was transmitted directly from birds to humans with lethal outcomes. Despite monumental efforts to contain them, the H5N1 viruses expanded their territory and caused a major outbreak in wild waterfowl in China in 2005. Indeed, they have even been transmitted to Siberia and Kazakhstan.
Despite extensive, coordinated efforts by various agencies and disciplines, both national and international, we are ill-equipped for a new influenza pandemic. In fact it is highly unlikely that adequate supplies of vaccine for the H5N1 viruses will be prepared prior to the occurrence of the next pandemic. Many countries are stockpiling influenza drugs, with the hope that the inevitable emergence of drug-resistant viruses will not nullify those efforts immediately. To combat the outbreaks that will undoubtedly occur in the near future a better understanding of influenza virus itself, the virus-host interaction, and mechanisms of drug resistance is urgently needed.
In this timely book world renowned scientists (including the 1996 Nobel Prize Winner, Peter Doherty) critically review the most important issues in this rapidly expanding field. Topics covered include analysis of influenza RNP, viral entry and intracellular transport, epidemiology, host range and pathogenicity, antivirals, vaccines, H5 viruses, and much more. Essential reading for all influenza virologists, molecular biologists, public health scientists and research scientists in pharmaceutical companies.
Reviews
"This book is highly recommended for infectious diseases specialists, virologists, vaccinologists, vaccine and drug development professionals, and anyone who wishes to learn more about influenza" from Clin. Inf. Diseases (2006) 43: 802-804.
"... interesting and readable ... lucidly written ... an important resource ..." from Clin. Inf. Diseases (2006) 43: 802-804.
"This is a wonderful book ... I have seldom read such a high-powered series of articles on molecular virology ... I do not think that the book will be easily outdated or even outclassed ..." from J. Antimicrobial Chemother. (2006) 58: 909.
"... excellent value for money and strongly recommended." from Microbiology Today (2006).
"should be on the shelf of every flu virologist, public health scientist, and vaccinologist ... I recommend it for all virologists and public health scientists who want to have the most updated picture on influenza and why a flu pandemic may occur in the near future. " from ASM Microbe (March 2007).
"This is one of the best books currently available on Influenza virus research." from Sai Vikram Vemula, Purdue University.
Table of contents
1. Structure and Function of the Influenza Virus RNP
Debra Elton, Paul Digard, Laurence Tiley and Juan Ortin
Influenza viruses have negative sense segmented RNA genomes, which are packaged into transcriptionally active ribonucleoproteins (RNPs). These RNPs are transcribed and replicated in the nucleus of host cells. During the replication cycle two types of positive sense RNA are synthesized; capped and polyadenylated messenger RNA and uncapped full length complementary (c)RNA. Complementary RNA acts as the replicative intermediate for synthesis of further negative sense genomic RNA. This cycle is carried out by the viral RNA-dependent RNA polymerase, a heterotrimeric complex which binds RNA through structure and sequence-specific interactions and has multiple functions including capped-RNA-binding activity, RNA endonuclease, polymerase and polyadenylation activities. These activities have specific roles during the viral transcription cycle and are controlled by interactions between the protein components and the RNA promoter structure. The mechanisms involved in the synthesis of viral messenger RNA are fairly well characterised, but less is known about the process of genome replication and the factors that control it. On the other hand, recent advances have been made towards elucidating the structure of the molecular machines responsible for virus RNA synthesis.
2. Entry and Intracellular Transport of Influenza Virus
Gary R. Whittaker and Paul Digard
All viruses need to recognize and enter target cells in order to cause infection. For the influenza viruses, an initial interaction with cell surface carbohydrate is followed by receptor-mediated endocytosis that traffics the virion into the endosomal pathway. Exposure to low pH in maturing endosomes triggers fusion of viral and cellular membranes leading to cytoplasmic uncoating of the virion. The released viral genomic ribonucleoproteins (RNPs) are imported into the nucleus where they are transcribed and replicated. Progeny RNPs are later exported to the cytoplasm and eventually arrive at the apical plasma membrane, where final virus assembly and budding take place. As with all viruses, infection is cyclical, and several important events occur during virus assembly and release that have profound effects on the entry process. In this review we survey both virus entry and intracellular transport of the viral components, highlighting both recent discoveries and the interdependence of virus assembly and entry. Where appropriate, we also highlight differences between members of the Orthomyxovirus family.
3. The Proton Selective Ion Channels of Influenza A and B Viruses
Robert A. Lamb and Lawrence H. Pinto
Influenza A and B viruses each encode via very different coding strategies a small oligomeric integral membrane protein, M2 of influenza A virus and BM2 of influenza B virus, and each protein is a proton selective ion channel. M2 and BM2 proteins have very different amino acid sequences but they share two key amino acid residues in the channel pore, a histidine and a tryptophan. These two residues provide a model of elegant simplicity for ionic selectivity and gating of these minimalistic ion channels. The activity of the ion channels are required during virus uncoating in the acidic environment of the endosome, to permit acidification of the interior of the virion particle which brings about protein-protein dissociation. The ion channels also equilibrate the acidic pH of the lumen of the trans Golgi network with the cytoplasm, during their own transport through the exocytic pathway. The influenza A virus M2 ion channel protein is the target of the antiviral drug amantadine ad the drug blocks directly ion channel activity. Thus, once the atomic structures of the M2 and BM2 ion channel proteins are known, it makes the channels attractive targets for rational drug design. The M2 and BM2 ion channel proteins may be multifunctional as the available data suggests the M2 cytoplasmic tail is involved in influenza virus assembly.
4. Receptor Specificity, Host-Range, and Pathogenicity of Influenza Viruses
Mikhail N. Matrosovich, Hans-Dieter Klenk and Yoshihiro Kawaoka
Influenza viruses attach to target cells via multivalent interactions of the viral hemagglutinin protein with sialyloligosaccharide moieties of cellular glycoconjugates. The interactions between the virus and cellular receptors and extracellular inhibitors determine virus host-range and tissue tropism. Sialic acids are ubiquitous on the surface of most avian and mammalian cells. Therefore, in addition to mediating infection of susceptible cells, influenza viruses can bind to a variety of other cell types leading to significant biological responses, such as polyclonal activation of B-lymphocytes, deactivation of neutrophils, and stimulation of inflammatory responses. Here, we discuss current knowledge of the influenza virus interactions with cellular receptors at the molecular level, outline methods used to characterize receptor specificity of influenza viruses, and give an overview of available data on the role of virus receptor specificity in host range restriction, interspecies transmission, and pathogenicity.
5. Dendritic Cells: Induction and Regulation of the Adaptive Immune Response to Influenza Virus Infection
Kevin L. Legge and Thomas J. Braciale
Clearance of respiratory viruses, like influenza virus, from the respiratory tract requires induction of an adaptive immune response. Initiation of adaptive immunity to foreign pathogens, like influenza virus, is thought to be mediated by dendritic cells. Dendritic cells perform this function by first sensing the invader in peripheral sites, maturing, and then migrating to the draining regional lymph nodes where they interact with and activate na�ve T and B cells. In this review, we will highlight and discuss what role dendritic cells may play in the induction and regulation of the adaptive immune response to pulmonary influenza virus infections, and how the interaction of influenza virus with dendritic cells may influence the shape of the developing adaptive immune response.
6. Quantitative and Qualitative Characterization of the CD8+ T cell Response to Influenza Virus Infection
Nicole L. La Gruta, Peter C. Doherty1 and 2
The CD8+ T cell response to influenza virus infection is critical for the efficient clearance of viral infection. The advent of tetramer staining has, over the last 5-10 years, enabled accurate quantitation of the epitope specific CD8+ T cell response to influenza virus infection, and has revealed much about immunodominance hierarchies and the kinetics of individual epitope specific responses. More recently, particular interest has been paid to the quality of CD8+ T cell responses, such as cytokine production and cytolytic ability, since T cell function must be a key factor in determining the efficacy of the response. Here, we describe recent advances in the characterization of both magnitude and quality of the CD8+ T cell response to influenza virus infection. These studies may also serve as a model to elucidate general mechanisms of CD8+ T cell-mediated viral clearance.
7. M2 and Neuraminidase Inhibitors: Anti-Influenza Activity, Mechanisms of Resistance, and Clinical Effectiveness
Larisa Gubareva, and Frederick G. Hayden
Antivirals have an important role in the treatment and prevention of influenza infections. This chapter describes the antiviral activity, mechanisms of action and resistance, clinical efficacy, and consequences of antiviral resistance for two available classes of anti-influenza drugs. Amantadine and rimantadine target the M2 protein of influenza A viruses; single mutations in the trans-membrane domain of M2 confer high-level resistance to this drug class. Therapeutic use is frequently associated with emergence of drug-resistant variants; such variants are transmissible from person-to-person and pathogenic. The approved neuraminidase inhibitors (zanamivir and oseltamivir) and investigative drugs are potent, specific inhibitors of influenza A and B viruses. Resistance emerges in vitro due to point mutations in hemagglutinin that alter cellular receptor binding or in viral neuraminidase that alter drug binding. Zanamivir and oseltamivir are highly effective for prophylaxis of influenza A and B infections; early therapeutic use reduces illness duration, lower respiratory complications, and in the case of oseltamivir, hospitalizations. Resistant variants with neuraminidase mutations have be infrequently isolated from adults and more commonly from pediatric patients treated with oseltamivir. Available evidence indicates that the relative efficiency of resistance emergence, the biologic fitness of resistant variants, and their transmissibility varies for two drug classes and for specific drugs within the neuraminidase inhibitor class. These differences have important implications for their clinical use.
8. Influenza Vaccines: Current and Future Strategies
Jacqueline M. Katz, Sanjay Garg, and Suryaprakash Sambhara
Vaccination is the primary method for the prevention of influenza and its complications. The continual genetic and antigenic variation that influenza viruses undergo requires constant global surveillance to identify and select new variants with epidemic potential or novel viruses with pandemic potential for inclusion in vaccines. Two general types of influenza vaccines, inactivated or live attenuated vaccines, both grown in embryonated hen's eggs, are currently licensed for use. Inactivated vaccines induce immunity to infection in 70-90% of healthy adults < 65 years of age when there is a good antigenic match between vaccine and circulating virus strain, but are generally less effective in older adults. Improved vaccines against epidemic influenza and effective vaccines against potential pandemic viruses are a public health priority. New strategies for influenza vaccines include altering the dose, site, or method of delivery of inactivated vaccines, the use of adjuvants or immunomodulators to enhance immune responses, or targeting viral proteins that may promote broader, cross-protective responses. Plasmid-based reverse genetics technology may provide a more rapid approach to the generation of candidate vaccine strains, and is essential for vaccine strains derived from highly pathogenic avian viruses. Cell culture-based vaccines may improve manufacturing capacity, particularly in the event of a newly emergent pandemic threat.
9. Epidemiology and Control of Human and Animal Influenza
Kanta Subbarao, David Swayne, and Christopher W. Olsen
Influenza viruses are clinically and economically important agents of disease in people, horses, pigs, marine mammals and poultry. Human influenza results from infection with influenza A, B or C viruses and a wide variety of domestic and free-ranging wild animal species can be infected with influenza A viruses. Aquatic birds are the natural hosts of influenza A viruses and represent a vast, global reservoir of influenza genes. Because pandemic influenza is fundamentally a zoonotic disease involving interspecies transmission of viruses from animals, this chapter jointly reviews the epidemiology, ecology and evolution of influenza viruses among humans, birds, and pigs. The epidemiologic consequences of genetic reassortment and adaptation of influenza viruses in these species and interspecies transmission are discussed. The epidemiology of interpandemic human influenza is presented with an emphasis on events of the past decade.
10. H5 Influenza Viruses
Robert G. Webster
Influenza viruses are classified into 15 subtypes on the basis of the hemagglutinin (HA) that they carry. Two subtypes found in aquatic birds worldwide-the H5 and H7 subtypes-are unique in having the ability to become highly pathogenic for domestic poultry and, occasionally, for humans after interspecies transmission. This chapter will primarily describe H5 influenza viruses. After transfer from the reservoir in aquatic birds to other avian species, H5 viruses can undergo rapid evolution: they may acquire multiple basic amino acids in the connecting peptide of the HA cleavage site, lose carbohydrate-bearing residues from the HA, undergo shortening of the neuraminidase (NA) stalk length, and acquire mutations in multiple genes encoding internal proteins including the polymerase PB2 and the nonstructural (NS) protein. Each of these events and the ecological conditions promoting their occurrence are considered in this chapter.
11. The Origin and Virulence of the 1918 'Spanish' Influenza Virus
Jeffery K. Taubenberger and Peter Palese
The 'Spanish' influenza pandemic of 1918-1919 caused acute illness in 25-30% of the world's population and resulted in the death of up to an estimated 40 million people. Using fixed and frozen lung tissue of 1918 influenza victims, the complete genomic sequence of the 1918 influenza virus is being deduced. Sequence and phylogenetic analysis of the completed 1918 influenza virus genes shows them to be the most avian-like among the mammalian-adapted viruses. This finding supports the hypothesis that (1) the pandemic virus contains genes derived from avian-like influenza virus strains and that (2) the 1918 virus is the common ancestor of human and classical swine H1N1 influenza viruses. The relationship of the 1918 virus with avian and swine influenza viruses is further supported by recent work in which the 1918 hemagglutinin (HA) protein crystal structure was resolved. Neither the 1918 hemagglutinin (HA) nor the neuraminidase (NA) genes possess mutations known to increase tissue tropicity that account for virulence of other influenza virus strains like A/WSN/33 or the highly pathogenic avian influenza H5 or H7 viruses. Using reverse genetics approaches, influenza virus constructs containing the 1918 HA and NA on an A/WSN/33 virus background were lethal in mice. The genotypic basis of this virulence has not yet been elucidated. The complete sequence of the non-structural (NS) gene segment of the 1918 virus was deduced and also tested for the hypothesis that enhanced virulence in 1918 could have been due to type I interferon inhibition by the NS1 protein. Results from these experiments suggest that in human cells the 1918 NS1 is a very effective interferon antagonist, but the 1918 NS1 gene does not have the amino acid change that correlates with virulence in the H5N1 virus strains identified in 1997 in Hong Kong. Sequence analysis of the 1918 pandemic influenza virus is allowing us to test hypotheses as to the origin and virulence of this strain. This information should help elucidate how pandemic influenza virus strains emerge and what genetic features contribute to virulence in humans.
12. Signaling and Apoptosis in Influenza Virus-Infected Cells
Stephan Ludwig
Infection of cells with viruses commonly leads to the activation of a variety of different signaling pathways within the infected cells. It is quite obvious that DNA viruses or retroviruses with DNA intermediates in their replication cycle need these signaling processes to activate the cellular DNA and protein synthesis machinery. However, it is not so clear how RNA viruses, such as influenza viruses cope with intracellular signaling. Since intracellular signaling pathways represent key switches in the determination of cell fate, the knowledge about their activation and function in infected cells may help to unravel some of the secrets of virus-host cell interactions. In this overview we will focus on recent advances on the function of intracellular signaling pathways and the induction of the apoptotic program in influenza virus infected cells. The role of these signaling events in the constant struggle between efficient virus propagation and the innate antiviral defense will be discussed.
13. Insights into Influenza Virus-Host Interactions Through Global Gene Expression Profiling: Cell Culture Systems to Animal Models
Marcus J. Korth, John C. Kash, Carole R. Baskin, and Michael G. Katze
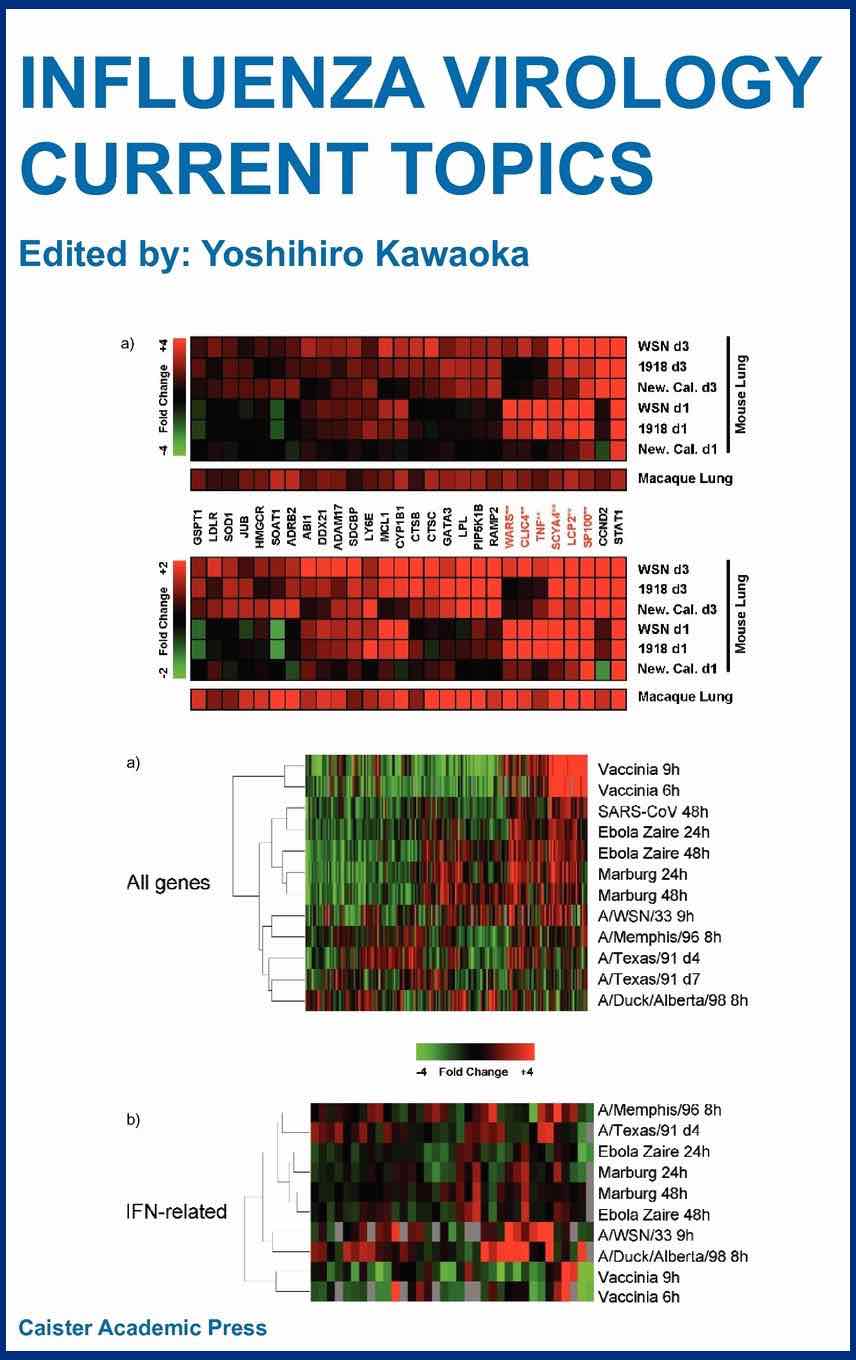
Researchers attempting to study a biological process as complex as the host response to a pathogen have long been faced with difficult choices. Simple model systems, such as cultured cell lines, provide considerable control over experimental variables, and methods that focus on tightly defined parameters can yield results that are often readily interpretable. But simple systems may not be representative of a natural infection, and data generated by focusing on a single gene, protein, or pathway can be difficult to integrate into a global picture. Today, genomic technologies such as DNA microarrays make it possible to perform experiments that provide a near comprehensive view of even such intricate processes as pathogen-host interactions. Still, choosing an experimental infection system remains difficult, and data interpretation has never been more complicated. In this chapter, we describe how gene expression profiling is being used to examine the host response to influenza virus. We discuss how the application of this technology has progressed from simple experimental systems, such as the in vitro infection of HeLa cells, to highly complex systems such as mouse and nonhuman primate models of infection. We also discuss the challenges associated with interpreting huge volumes of data generated from microarray analyses, and describe how comparing the host's transcriptional response to infection by diverse wild type or engineered viruses is providing new insights into the characteristics of influenza virus that contribute to its variable virulence.
How to buy this book
(EAN: 9781904455066 9781913652319 Subjects: [virology] [microbiology] [medical microbiology] )