Abstract
Objectives
CT findings of COVID-19 look similar to other atypical and viral (non-COVID-19) pneumonia diseases. This study proposes a clinical computer-aided diagnosis (CAD) system using CT features to automatically discriminate COVID-19 from non-COVID-19 pneumonia patients.
Methods
Overall, 612 patients (306 COVID-19 and 306 non-COVID-19 pneumonia) were recruited. Twenty radiological features were extracted from CT images to evaluate the pattern, location, and distribution of lesions of patients in both groups. All significant CT features were fed in five classifiers namely decision tree, K-nearest neighbor, naïve Bayes, support vector machine, and ensemble to evaluate the best performing CAD system in classifying COVID-19 and non-COVID-19 cases.
Results
Location and distribution pattern of involvement, number of the lesion, ground-glass opacity (GGO) and crazy-paving, consolidation, reticular, bronchial wall thickening, nodule, air bronchogram, cavity, pleural effusion, pleural thickening, and lymphadenopathy are the significant features to classify COVID-19 from non-COVID-19 groups. Our proposed CAD system obtained the sensitivity, specificity, and accuracy of 0.965, 93.54%, 90.32%, and 91.94%, respectively, using ensemble (COVIDiag) classifier.
Conclusions
This study proposed a COVIDiag model obtained promising results using CT radiological routine features. It can be considered an adjunct tool by the radiologists during the current COVID-19 pandemic to make an accurate diagnosis.
Key Points
• Location and distribution of involvement, number of lesions, GGO and crazy-paving, consolidation, reticular, bronchial wall thickening, nodule, air bronchogram, cavity, pleural effusion, pleural thickening, and lymphadenopathy are the significant features between COVID-19 from non-COVID-19 groups.
• The proposed CAD system, COVIDiag, could diagnose COVID-19 pneumonia cases with an AUC of 0.965 (sensitivity = 93.54%; specificity = 90.32%; and accuracy = 91.94%).
• The AUC, sensitivity, specificity, and accuracy obtained by radiologist diagnosis are 0.879, 87.10%, 88.71%, and 87.90%, respectively.
Similar content being viewed by others
Introduction
In December 2019, a novel coronavirus-infected pneumonia, called coronavirus disease 2019 (COVID-19), occurred in the city of Wuhan, China, related to Huanan Seafood Market [1,2,3]. This outbreak has spread exponentially throughout the world and is declared a pandemic [4]. The most prevalent clinical symptoms of COVID-19 patients are fever, followed by cough, fatigue, and dyspnea. It can lead to acute respiratory distress syndrome, acute renal failure, shock, and death [3, 5].
The diagnostic criteria of COVID-19 pneumonia are laboratory evaluation of respiratory secretions acquired from endotracheal aspirate, bronchoalveolar lavage, or nasopharyngeal/ oropharyngeal swab [6]. Currently, laboratory examination such as reverse transcriptase-polymerase chain reaction (RT-PCR) test has become the standard assessment for the diagnosis of COVID-19 infection [7, 8]. However, RT-PCR testing results may be falsely negative due to insufficient specimen or laboratory error [9]. In addition, although the image finding can be positive in the early stages of the disease, RT-PCR results can be negative at the early stages in some cases. However, RT-PCR can become positive in the following course of the disease [10, 11]. Therefore, a combination of repeated swab tests and CT imaging can be used as a tool to diagnose the individual with negative RT-PCR screening and high suspicion of COVID-19 infection [10].
Chest CT scan provides more detailed information about the chest and hence, it is used to diagnose COVID-19 patients. In a study using 1014 patients, the sensitivity of chest CT in suggesting COVID-19 based on the positive RT-PCR is 97%, and patients with negative RT-PCR and chest CT findings of 75% are positive [12]. Abnormal CT findings such as pneumonia, the existence of patterns like ground-glass opacity (GGO), and bilateral patchy shadowing are frequently observed in positive COVID-19 cases [13]. The most frequent CT features of COVID-19 pneumonia are GGO, crazy-paving pattern, mixed GGO and consolidation, bilateral lobe involvement, and subpleural lesions [14, 15].
Radiologists can help in several ways in this current pandemic such as (i) early detection of the disease and plan ahead for proper management in later stages of the disease; (ii) score the severity of the disease and help to identify the chance of developing ARDS and the need to transferring to the intensive care unit; and (iii) detect possible secondary or co-infection of bacterial pneumonia, which is very critical as bacterial pneumonia can lead to serious complications [16]. However, both COVID-19 virus and other non-COVID-19 viruses can cause pneumonia and differentiate them, which is challenging for radiologists as both CT findings look similar [15, 17]. Bai et al [18] showed that seven radiologists can diagnose COVID-19 pneumonia with mean sensitivity and specificity of 70.42% and 83.71%, respectively. Also, they concluded that the radiologists showed high specificity but moderate sensitivity in distinguishing COVID-19 pneumonia from other atypical and viral (non-COVID-19) pneumonia based on chest CT findings. To overcome these limitations and manage the COVID-19 pneumonia patients effectively, a computer-aided diagnosis (CAD) system is needed [19].
Nowadays, CAD systems can help and allow radiologists to make a better decision, especially in CT lung imaging [20,21,22]. It also help to detect lung abnormalities [23, 24] and pulmonary fibrosis [25, 26], manage lung nodules [27, 28], and differentiate nodules from interferential vessels [29, 30]. In this work, we have investigated the potential of using the CAD system to diagnose and manage patients with COVID-19 pneumonia disease. In this work, we proposed a clinical CAD system, namely COVIDiag, to differentiate COVID-19 from non-COVID-19 pneumonia diseases using features extracted from the chest CT images. We feel that the proposed system can help to reduce the workload and improve the quality of COVID-19 disease diagnosis.
Patients and methods
Patients
Regardless of demographic values like age and gender in the pandemic of the COVID-19, the patients with flu-like symptoms and diagnosed with novel coronavirus were enrolled for the study. A chest high-resolution CT (HRCT) examination was conducted for all patients before enrolling them in this study. The confirmation for COVID-19 was done through RT-PCR based on nasopharyngeal swab samples. The patients with respiratory infections with negative RT-PCR and confirmed laboratory test were excluded in this study. Also, those cases with chronic lung diseases and subsequent pulmonary involvement were excluded. HRCT images of patients with other causes of atypical and viral pneumonia, such as adenoviral or H1N1 flu from PACS of our university hospital, were retrospectively investigated from January 2018 to December 2019.
Image acquisition
All HRCT examinations were performed using a 16-MDCT scanner (Alexion, Canon Medical Systems) with high-resolution protocol: patients in the recumbent situation with the arms over the head; 1- to 2-mm slice thickness in increments of up to 10 mm from the lung apices through the hemidiaphragm, at deep inspiration; tube voltage, 120 kVp; tube current time, 50–100 mAs; and pitch, 0.8–1.5. Parenchymal window settings were set for all patients to a range of window level and a window width of − 600 to − 500 Hounsfield units (HU) and 1500 to 1600 HU, respectively. All of the CT slices were reconstructed using an iterative algorithm (adaptive iterative dose reduction using three-dimensional processing (AIDR 3D)) with the kernel FC56, and scans were acquired without the use of contrast agent.
CT feature extraction
Few studies indicated that the pattern, location, and distribution of lesions can differentiate COVID-19 from other non-COVID-19 pneumonia [14, 18, 31]. There are few radiological features such as GGO, crazy-paving, peripheral, both peripheral and central involvement, and upper lobe involvement, which are more common in COVID-19 pneumonia compared with non-COVID-19 pneumonia. On the other hand, there are few other radiological features that are more common and specific in non-COVID-19 pneumonia compare with that in COVID-19 pneumonia, such as pleural effusion, pleural thickening, air bronchogram, consolidation, central involvement, and lymphadenopathy.
In this study, two radiologists with more than 15 years of experience in thoracic imaging, who were blinded to the laboratory test, reviewed the CT images. Radiological features were extracted by one radiologist and confirmed by another experienced radiologist. In total, 20 radiological features are extracted for both the groups. These radiological features are as follows:
- (a)
Location 1, location of lesion(s) are evaluated if they involve unilateral, bilateral, or both unilateral and bilateral;
- (b)
Location 2, location of lesion(s) are studied if they are present in lower, upper, or both lobes;
- (c)
Distribution of lesion(s) are defined as peripheral, central, or both central and peripheral;
- (d)
Number lesion(s) is assigned as a single lesion, if there is only one patch of a lesion, multiple lesions, if there are 2–4 patches of lesions in every lung, and diffuse lesion, if lesions involved the entire lobe bilaterally;
- (e)
GGO, which is hazy augmented lung attenuation with the maintenance of bronchial and vascular borders. In other words, a hazy opacity that does not obscure the underlying pulmonary vessel;
- (f)
Consolidation, which is described as opacification with obscuration of vessels and airway borders walls. It is defined as filling of air that usually fills the small airways with something else;
- (g)
Presence of reticular: every thin linear opacity between 1 and 3 mm thickness;
- (h)
Nodule, which is defined as every round or oval well-defined margin opacity;
- (i)
Vascular thickening;
- (j)
Septal thickening;
- (k)
Bronchial wall thickening;
- (l)
Air bronchogram; which is defined as opacification of surrounding alveoli (gray/white) make the air-filled bronchi (dark) detectable;
- (m)
Cavity;
- (n)
Cyst;
- (o)
Crazy-paving, which is a linear pattern superimposed on an area of GGO, with irregular paving stones pattern;
- (p)
Halo sign;
- (q)
Reversed halo sign;
- (r)
Pleural effusion, defined as blunting of the costophrenic angle, cardiophrenic angle, and fluid within the horizontal or oblique fissures;
- (s)
Pleural thickening; and
- (t)
Lymphadenopathy, described as a lymph node with a greater size than 1 cm in short axis.
Machine learning study
The MATLAB software (version R2019b, MathWorks Inc) was used to implement machine-learning process. In order to perform an automated diagnosis of COVID-19 cases, five classifiers are used: decision tree, K-nearest neighbor (KNN), 3- naïve Bayes, support vector machine (SVM), and ensemble. The optimization method based on the Bayesian optimization algorithm [32] is used to define the optimized hyperparameters. This method searches the specific hyperparameters within their ranges for each classifier to find the bestpoint hyperparameters to yield the highest classification performance. The names of hyperparameters and their ranges (in parentheses) for each classifier are as follows: decision tree: maximum number of splits (1–487), split criterion (Gini’s diversity index, maximum deviance reduction); KNN: number of neighbors (1–244), distance metric (city block, Chebyshev, correlation, cosine, Euclidean, Minkowski, Mahalanobis, spearman, hamming, and Jaccard), distance weight (equal, inverse, squared inverse); naïve Bayes: distribution name (Gaussian, kernel), kernel type (Gaussian, box, Epanechnikov, triangle); SVM: kernel function (Gaussian, linear, quadratic, cubic), kernel scale (0.001–1000) and box constraint level (0.001 to 1000); and Ensemble: ensemble method (AdaBoost, RUSBoost, LogitBoost, GentleBoost, and bag), maximum number of splits (1–487), number of learners (10–500), learning rate (0.001–1).
In this study, the entire database is divided into two parts: 80% for training and 20% for testing. All five classifiers are trained for 30 iterations using the Bayesian optimization algorithm. The K-fold (K = 20) cross-validation strategy is used to prevent over-fitting of the models. At the end of the training process, optimization algorithm returns the bestpoint hyperparameters for each classifier.
Statistical analysis
The discrimination between COVID-19 and non-COVID-19 groups of CT features is evaluated with the chi-square test. Statistically significant features have a p value of less than 0.05.
Performance evaluation of networks
Five parameters namely sensitivity, specificity, accuracy, PPV, and NPV are calculated in our study to compare the performance of radiologists and classifiers. COVID-19 and other viral pneumonia (non-COVID-19 group) cases are considered positive and negative, respectively. Therefore, correctly diagnosed COVID-19 and non-COVID-19 cases are indicated as NTP and NTN, respectively. Also, incorrectly diagnosed COVID-19 and non-COVID-19 cases are identified as NFP and NFN, respectively. Furthermore, ROC curve analysis is used and AUC is computed [33]. SPSS software (version 24.0, IBM Corporation) is used for statistical analysis. Figure 1 shows the steps involved in our study at a glance.
Results
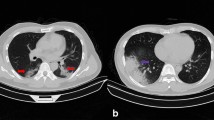
In this study, 612 patients (306 COVID-19 and 306 non-COVID-19) were recruited. In total, 488 patients (with 50–50 distribution) were used for the training phase and the rest of the patients (20%) were used to test the developed model. Figure 2 shows the sample CT images of patients with COVID-19 and non-COVID-19 pneumonia.
CT sample images of patients with pneumonia. a A 28-year-old male with confirmed COVID-19 pneumonia. The red arrow in the right upper lobe indicates mixed ground glass and crazy paving opacity. b A 67-year-old female patient with confirmed COVID-19 pneumonia. The red arrows indicate multifocal ground-glass opacity pattern in both lobes. c An 68-year-old male patient with atypical pneumonia. The red arrows indicate mixed ground glass and alveolar consolidation pattern in the right lower lobe. d A 67-year-old male patient with H1N1 pneumonia. The red and yellow arrows indicate alveolar consolidation the right and left upper lobe, respectively
CT findings
The bilateral involvement is significantly high in COVID-19 patients (176 out of 244, 72.13%) compared with that in the non-COVID-19 group (72 out of 244, 29.5%). In the location 2 feature, the infection involvement of the upper, lower, and both lobes in COVID-19 group is observed in 106 (43.44%), 48 (19.67%), and 90 (36.89%) patients, respectively, which are significant differences compared with the non-COVID-19 group whose involvements are observed in 131 (53.69%), 89 (36.47%), and 24 (09.84%) cases, respectively. The peripheral, central, and both central and peripheral involvements in COVID-19 group are discovered in 147 (60.25%), 26 (10.65%), and 71 (29.10%) cases, respectively, for the distribution feature, which have shown significant differences compared with the non-COVID-19 group whose the involvements are observed in 41 (16.80%), 115 (47.13%), and 88 (36.07%) cases, respectively (Table 1).
The single, multiple, and diffuse lesions in the COVID-19 group are observed in 32 (13.11%), 136 (55.74%), and 76 (31.15%) cases, respectively, for the lesion feature compared with those in the non-COVID-19 group whose single, multiple, and diffuse lesions are found in 155 (63.52%), 74 (30.33%), and 15 (06.15%) cases, respectively, with p value < 0.001. In addition, the GGO and crazy-paving features are found significantly high in COVID-19 cases compared with those in the non-COVID-19 group (p < 0.001). In contrast, consolidation, reticular, bronchial wall thickening, nodule, air bronchogram, cavity, pleural effusion, pleural thickening, and lymphadenopathy are more common in the non-COVID-19 group. However, no significant differences are seen in other CT features like vascular thickening, septal thickening, cyst, halo sign, and reversed halo sign (Table 1).
Performance of machine learning and radiologist
The results of the optimization process and the hyperparameters of each optimized network are shown in Fig. 3.
The optimization curves of five networks after 30 iterations. a Decision tree; b K-nearest neighbor; c naïve Bayes; d support vector machine; and (e) ensemble (named as COVIDiag). During the process, the optimization algorithm seeks different combinations in each iteration to find the condition with the minimum classification error and confidence interval, i.e., “bestpoint hyperparameters”
The classification results of the models for COVID-19 and non-COVID-19 groups are summarized in Table 2. Also, we measured the performance of the radiologist as a baseline to compare the results with these five models. Among all models, the highest performance is obtained for ensemble classifier with an AUC of 0.988 (sensitivity, 94.67%; specificity, 93.03%; accuracy, 93.85%) for the training dataset. In contrast, the lowest performance is obtained for decision tree with an AUC of 0.934 (sensitivity, 89.34%; specificity, 90.16%; accuracy, 89.75%). After training the models, they are tested with blinded (unseen) data. Then, the highest discriminative power is obtained for the ensemble model with AUC, sensitivity, specificity, and accuracy of 0.965, 93.54%, 90.32%, and 91.94%, respectively. Also, the AUC, sensitivity, specificity, and accuracy obtained for the diagnosis by a radiologist are 0.879, 87.10%, 88.71%, and 87.90%, respectively (Table 2). Radar plots and ROC curves for various classifiers and radiologist in the testing phase are presented in Fig. 4a and b, respectively. The COVIDiag model is available (Link) (to test the model with your own data, please follow the guide sheet (Figure E1, Supplementary material).
Discussion
In this study, the best performance is achieved by ensemble classifier (COVIDiag) with an AUC of 0.965. The main advantage of this classifier is that it uses many (81 in this study) learners to build an accurate model. Aggregating the output of the learners help to build a robust model compared to the individual learner [34]. Hence, the stability and discriminative power of the ensemble classifier is higher than other classifiers used in this study. In addition, our results indicate that the performance of the COVIDiag is even higher than the radiologist for the testing dataset (AUC of COVIDiag vs. radiologist: 0.965 vs. 0.879). In this work, 58 out of 62 COVID-19 cases and 56 out of 62 non-COVID-19 cases are correctly diagnosed by COVIDiag. The details about the diagnosis results of COVIDiag and radiologists are listed in Tables E1 and E2 (supplementary material). The machine-learning model can deal with complex and multiparametric data better than the radiologists. During the visual inspection process, radiologists should extract several radiological features from the CT images, make a meaningful relationship between them, and finally make a final decision. This step of processing is subjective, time-consuming, and prone to human errors. Hence, in this study, we proposed a practical clinical CAD system (COVIDiag) to help radiologists during routine practices.
The results of the present study indicate that multiple and diffused lesions with GGO and crazy-paving patterns are significantly more common in COVID-19 pneumonia cases. On the other hand, patients with cavity, nodule, single lesion, consolidation, reticular, bronchial wall thickening, air bronchogram, pleural effusion, pleural thickening, and lymphadenopathy are significantly more likely to have non-COVID-19 pneumonia. Moreover, the bilateral involvements with peripheral distribution occur more significantly in patients with COVID-19 pneumonia. Our findings in terms of distribution, GGO, pleural effusion, and pleural thickening are similar, but the terms of nodule, location 1, bronchial wall thickening, crazy-paving, halo, reverse halo, and vascular thickening are not similar to the study by Bai et al [18]. In addition, except for GGO, lymphadenopathy, and pleural effusion, the results of other features are similar to those of Long et al [31] and Cheng et al [35]. The main reason for the difference in the results is that these studies have used either a small number of patients in both groups or included all types of non-COVID-19 pneumonia patients. Hence, bacterial or other atypical pneumonia cases may be included. On the other hand, during the diagnosing process, radiologists should pay attention to the time of patient’s admission (Table 3) and the similar CT findings that can be misdiagnosed as COVID-19 pneumonia (Table 4) to reduce the possible false-positive cases.
Few artificial intelligence studies on chest CT images have been emerging to help physicians to manage patients with COVID-19 pneumonia. Some studies reported that deep learning could diagnose COVID-19 pneumonia cases with an AUC of 0.960 [36] and 0.994 [37], respectively. However, we used simple machine-learning technique and achieved an AUC of 0.965. Hence, the COVIDiag is more effective in discriminating COVID-19 pneumonia cases from non-COVID-19 cases. The main advantage of the proposed model is simple and takes less time to train as it is not deep learning–based model.
Another advantage of the COVIDiag is that it is easy to use. After the acquisition of CT images from the patients, we can extract the desired features from the images and feed those features on the pre-trained model to get the output class. In addition, COVIDiag is reproducible and can be used for unlimited time in a day without degrading the performance. These test images again can be used to train the model and make the model more robust. Also, this system is more economical and can be used along with the RT-PCR method. The RT-PCR method is expensive as it involved well-equipped laboratories which many underdeveloped countries may not be able to afford [38]. In addition, countries continuously need to supply the high demand for the new kits. In the present scenario, our proposed COVIDiag can be used to meet the challenges of third-world countries and help to rehabilitate the affected patients immediately by accurate faster diagnosis.
The limitation of our proposed system is that CT findings can be negative in the early stage, while the RT-PCR is positive [39, 40]. In this situation, the results of COVIDiag tend to be negative. Another limitation is that, in some cases, the initial results of RT-PCR may be false negative [9]. So, these patients with COVID-19 may be excluded incorrectly. It should be noted that the PPV and NPV indices are not intrinsic to the test and they depend on the prevalence of diseases. In this study, we provided balanced dataset, which can affect the indices.
Conclusion
This study proposed an automated clinical COVIDiag system based on routine radiological parameters and machine-learning techniques. The developed tool is simple to operate and can help the radiologists to reduce their daily workload by helping them to make an accurate diagnosis. In the future, we intend to extend our COVIDiag model to assess the severity of COVID-19 patients.
Abbreviations
- ARDS:
-
Acute respiratory distress syndrome
- CAD:
-
Computer-aided diagnosis
- COVID-19:
-
Coronavirus disease 2019
- GGO:
-
Ground-glass opacity
- HRCT:
-
High-resolution CT
- HU:
-
Hounsfield units
- KNN:
-
K-nearest neighbor
- RT-PCR:
-
Reverse transcriptase-polymerasechain reaction
- SVM:
-
Support vector machine
References
Lu H, Stratton CW, Tang Y-W (2020) Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol 92:401–402
Hui DS, Azhar EI, Madani TA et al (2020) The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health: the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91:264–266
Huang C, Wang Y, Li X et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506
World Health Organization (2020) WHO announces COVID-19 outbreak a pandemic. Available via http://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic. Accessed 3 June 2020
Yang X, Yu Y, Xu J et al (2020) Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8:475–481
Center for Disease Control and Prevention (2020) Interim guidelines for collecting, handling, and testing clinical specimens from persons under investigation (PUIs) for coronavirus disease 2019 (COVID-19). Available via https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed 3 June 2020
World Health Organization (WHO) (2020) Novel Coronavirus. (2019-nCoV) technical guidance: laboratory guidance. Geneva: WHO. Available via https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance. Accessed 3 June 2020
World Health Organization (2020) Clinical management of severe acute respiratory infection when COVID-19 is suspected: interim guidance. Available via https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected. Accessed 3 June 2020
FDA (2020) Fact sheet for healthcare providers: CDC - 2019-nCoV real-time RT-PCR diagnostic panel. Available via https://www.fda.gov/media/134920/download. Accessed 3 June 2020
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. https://doi.org/10.1148/radiol.2020200343
Huang P, Liu T, Huang L et al (2020) Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology 295:22–23
Ai T, Yang Z, Hou H et al (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. https://doi.org/10.1148/radiol.2020200642
W-j G, Ni Z-y HY et al (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720
Zu ZY, Jiang MD, Xu PP et al (2020) Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. https://doi.org/10.1148/radiol.2020200490
Chung M, Bernheim A, Mei X et al (2020) CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 295:202–207
Kim H (2020) Outbreak of novel coronavirus (COVID-19): what is the role of radiologists? Eur Radiol 30:3266–3267
Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH (2020) Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. Radiology. https://doi.org/10.1148/radiol.2020200527
Bai HX, Hsieh B, Xiong Z et al (2020) Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. https://doi.org/10.1148/radiol.2020200823
Marten K, Engelke C (2007) Computer-aided detection and automated CT volumetry of pulmonary nodules. Eur Radiol 17:888–901
Vassallo L, Traverso A, Agnello M et al (2019) A cloud-based computer-aided detection system improves identification of lung nodules on computed tomography scans of patients with extra-thoracic malignancies. Eur Radiol 29:144–152
Zhao Y, de Bock GH, Vliegenthart R et al (2012) Performance of computer-aided detection of pulmonary nodules in low-dose CT: comparison with double reading by nodule volume. Eur Radiol 22:2076–2084
Christe A, Leidolt L, Huber A et al (2013) Lung cancer screening with CT: evaluation of radiologists and different computer assisted detection software (CAD) as first and second readers for lung nodule detection at different dose levels. Eur J Radiol 82:e873–e878
Bhandary A, Prabhu GA, Rajinikanth V et al (2020) Deep-learning framework to detect lung abnormality – a study with chest X-ray and lung CT scan images. Pattern Recogn Lett 129:271–278
Liang G, Zheng L (2020) A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed 187:104964
Christe A, Peters AA, Drakopoulos D et al (2019) Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images. Invest Radiol 54:627–632
Walsh SLF, Calandriello L, Silva M, Sverzellati N (2018) Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med 6:837–845
Gong J, Liu J, Hao W et al (2020) A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on CT images. Eur Radiol 30:1847–1855
Le V, Yang D, Zhu Y et al (2018) Quantitative CT analysis of pulmonary nodules for lung adenocarcinoma risk classification based on an exponential weighted grey scale angular density distribution feature. Comput Methods Programs Biomed 160:141–151
Sun L, Wang Z, Pu H et al (2020) Spectral analysis for pulmonary nodule detection using the optimal fractional S-transform. Comput Biol Med 119:103675
Zhang W, Wang X, Li X, Chen J (2018) 3D skeletonization feature based computer-aided detection system for pulmonary nodules in CT datasets. Comput Biol Med 92:64–72
Long C, Xu H, Shen Q et al (2020) Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol 126:108961
Snoek J, Larochelle H, Adams RP (2012) Practical Bayesian optimization of machine learning algorithms. Adv Neural Inf Process Syst 25:2951–2959
van Erkel AR, Pattynama PMT (1998) Receiver operating characteristic (ROC) analysis: basic principles and applications in radiology. Eur J Radiol 27:88–94
Rokach L (2010) Ensemble-based classifiers. Artif Intell Rev 33:1–39
Cheng Z, Lu Y, Cao Q et al (2020) Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a single-center study in Shanghai, China. AJR Am J Roentgenol. https://doi.org/10.2214/AJR.20.22959
Li L, Qin L, Xu Z et al (2020) Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. https://doi.org/10.1148/radiol.2020200905
Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A (2020) Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med 121:103795
Bedford J, Enria D, Giesecke J et al (2020) COVID-19: towards controlling of a pandemic. Lancet 395:1015–1018
Bernheim A, Mei X, Huang M et al (2020) Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology 295:200463
Yang W, Cao Q, Qin L et al (2020) Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19): a multi-center study in Wenzhou city, Zhejiang, China. J Inf 80:388–393
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Afshin Mohammadi.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• cross-sectional study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 957 kb)
Rights and permissions
About this article
Cite this article
Abbasian Ardakani, A., Acharya, U.R., Habibollahi, S. et al. COVIDiag: a clinical CAD system to diagnose COVID-19 pneumonia based on CT findings. Eur Radiol 31, 121–130 (2021). https://doi.org/10.1007/s00330-020-07087-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07087-y